Phosphofructokinase I
Phosphofructokinase (PFK) is a key enzyme in glycolysis. It transfers a phosphate group to fructose and in doing so controls entry into the pathway. PFK is highly regulated and is cooperative for fructose and shows allosteric inhibition by phosphoenolpyruvate and activation by ADP. The enzyme changes from the R state to the T state by a rotation around the 'p' axis of the molecule, resulting in a change in the subunit-subunit interactions that are communicated to the active site.
The ATP binding site remains fixed during the transition from R to T state except for the residue arg 72 that is important for bridging the substrate phosphates during catalysis. In the T state this Arg residue forms a salt bridge with glu241. The T state is also characterised by a large change in the substrate binding site, the movement of the '6-F' loop leads to the collapse of this binding site.
Reference Protein and Structure
- Sequence
-
P0A796
 (2.7.1.11)
(2.7.1.11)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Escherichia coli K-12 (Bacteria)

- PDB
-
1pfk
- CRYSTAL STRUCTURE OF THE COMPLEX OF PHOSPHOFRUCTOKINASE FROM ESCHERICHIA COLI WITH ITS REACTION PRODUCTS
(2.4 Å)



- Catalytic CATH Domains
-
3.40.50.450
 3.40.50.460
3.40.50.460  (see all for 1pfk)
(see all for 1pfk)
- Cofactors
- Magnesium(2+) (1)
Enzyme Reaction (EC:2.7.1.11)
Enzyme Mechanism
Introduction
Asp128 is proposed to act as a base, abstracting a proton from 1-OH of 6-phosphofructose. This increases the nucleophilicity of the functional group which then attacks at the gamma phosphate of ATP in an SN2 reaction, forming 1,6-bisphosphate fructose and ADP.
Catalytic Residues Roles
| UniProt | PDB* (1pfk) | ||
| Asp104 | Asp103(104)A | Coordinates to the magnesium ion which in turn binds to the phosphate groups depending on the R or T state of the enzyme. | metal ligand |
| Asp130 | Asp129(130)A | Proximal to Asp128. Helps activate the general acid/base. | increase basicity, hydrogen bond acceptor, increase acidity |
| Thr126, Arg172, Arg73, Gly12 (main-N) | Thr125(126)A, Arg171(172)A, Arg72(73)A, Gly11(12)A (main-N) | Maintain a positively charged environment that stabilises the reactive intermediates formed during the course of the reaction. | electrostatic stabiliser |
| Asp128 | Asp127(128)A | Acts as a general acid/base. Proposed to abstract a proton from 1-OH of 6-phosphofructose. | activator, hydrogen bond acceptor, proton acceptor, proton donor |
Chemical Components
bimolecular nucleophilic addition, proton transfer, overall reactant used, overall product formed, rate-determining step, native state of enzyme regeneratedReferences
- Evans PR et al. (1981), Philos Trans R Soc Lond B Biol Sci, 293, 53-62. Phosphofructokinase: Structure and Control [and Discussion]. DOI:10.1098/rstb.1981.0059.
- Kloos M et al. (2015), Biochem J, 469, 421-432. Crystal structure of human platelet phosphofructokinase-1 locked in an activated conformation. DOI:10.1042/BJ20150251. PMID:26205495.
- Martinez-Oyanedel J et al. (2007), J Mol Biol, 366, 1185-1198. The first crystal structure of phosphofructokinase from a eukaryote: Trypanosoma brucei. DOI:10.1016/j.jmb.2006.10.019. PMID:17207816.
- Tlapak-Simmons VL et al. (1998), Biophys J, 75, 1010-1015. Obfuscation of allosteric structure-function relationships by enthalpy-entropy compensation. DOI:10.1016/S0006-3495(98)77589-7. PMID:9675201.
- LAINE R et al. (1992), Eur J Biochem, 207, 1109-1114. Interaction between the carboxyl groups of Asp127 and Asp129 in the active site of Escherichia coli phosphofructokinase. DOI:10.1111/j.1432-1033.1992.tb17148.x.
- Auzat I et al. (1992), Protein Sci, 1, 254-258. pH dependence of the reverse reaction catalyzed by phosphofructokinase I from Escherichia coli: Implications for the role of Asp 127. DOI:10.1002/pro.5560010207. PMID:1304907.
- Rypniewski WR et al. (1989), J Mol Biol, 207, 805-821. Crystal structure of unliganded phosphofructokinase from Escherichia coli. DOI:10.1016/0022-2836(89)90246-5. PMID:2527305.
- Shirakihara Y et al. (1988), J Mol Biol, 204, 973-994. Crystal structure of the complex of phosphofructokinase from Escherichia coli with its reaction products. DOI:10.1016/0022-2836(88)90056-3.
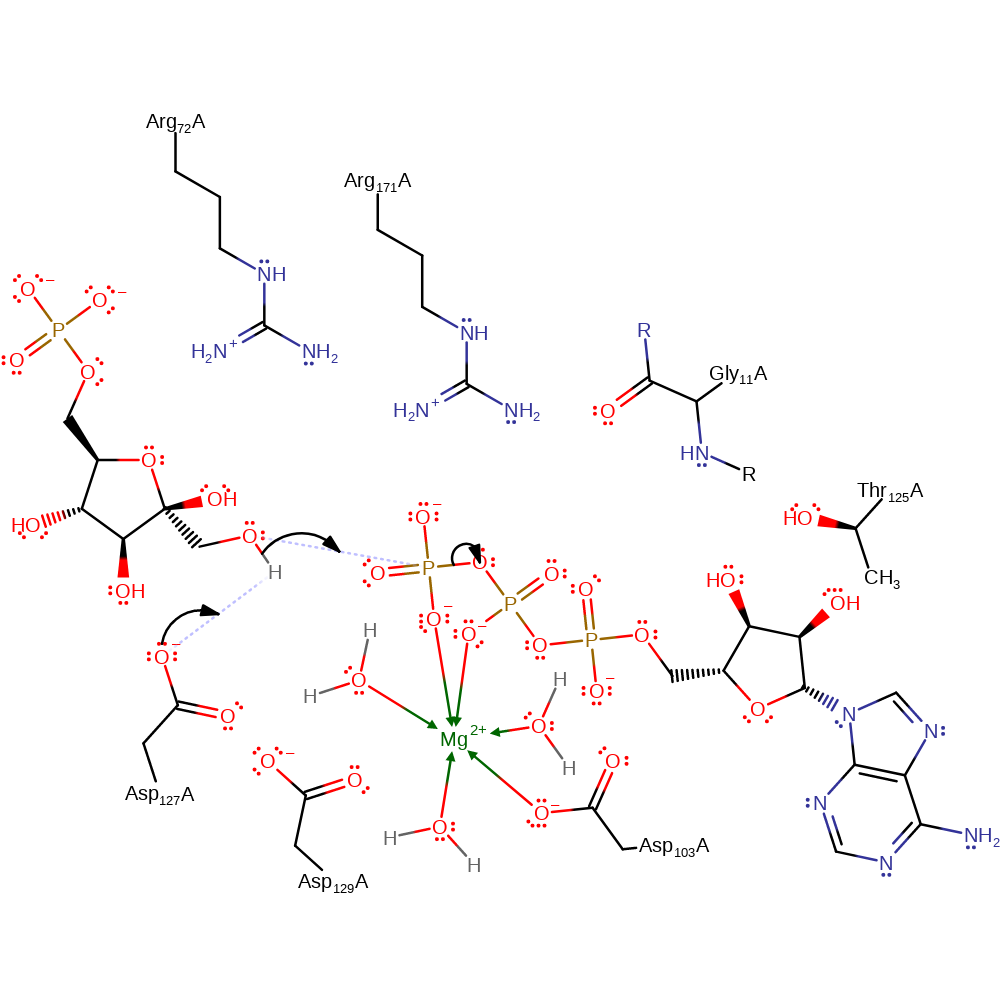
Step 1. 6-phosphofructose acts as an allosteric activator. On binding, 6-phosphofructose induces a reactive conformation which orientates both itself and ATP for base-catalysed phosphate transfer. 6-phosphofructose is activated as a nucleophile on deprotonation of the O-1 functional group by Asp128. The oxygen abstracts the gamma phosphate from ATP, forming ADP.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp127(128)A | activator, hydrogen bond acceptor |
| Asp129(130)A | hydrogen bond acceptor, increase basicity |
| Gly11(12)A (main-N) | hydrogen bond donor, electrostatic stabiliser |
| Arg72(73)A | electrostatic stabiliser |
| Arg171(172)A | electrostatic stabiliser |
| Thr125(126)A | electrostatic stabiliser |
| Asp103(104)A | metal ligand |
| Asp127(128)A | proton acceptor |
Chemical Components
ingold: bimolecular nucleophilic addition, proton transfer, overall reactant used, overall product formed, rate-determining step
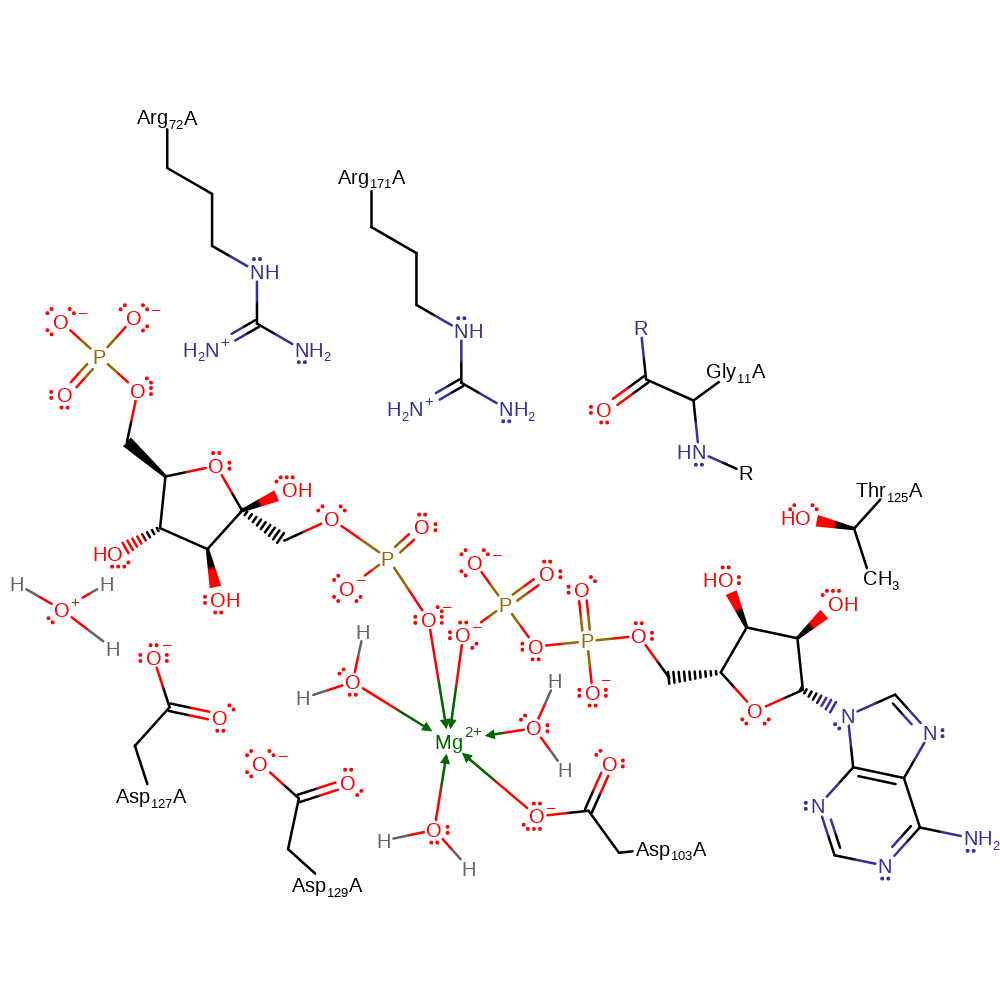
Step 2. Asp128 is deprotonated. Kinetic studies indicate that for the reverse reaction, Asp128 must be protonated. Crystal structures show a water molecule within the active site. This is thought to facilitate proton transfer from and to Asp128.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp129(130)A | increase acidity |
| Asp127(128)A | proton donor |





 Download:
Download: