DNA beta-glucosyltransferase
DNA beta-glucosyltransferase (BGT) is an enzyme encoded by a number of bacteriophage belonging to the T-even group. It catalyses the transfer of glucose from uridine diphosphoglucose (UDP-glucose) to 5-hydroxymethylcytosine (5-HMC) bases in double stranded DNA. Such glucosylation protects the infecting viral DNA from host restrictive enzymes. It may, in addition, be involved in phage specific gene expression by influencing transcription. Glucosylation also occurs in Trypanosoma bruceii where it is thought to be involved in regulating the expression of variant surface glycoproteins used by the parasite for protection against immune recognition. BGT is one of two enzymes involved in the glucosylation. In contrast to its counterpart which catalyses the formation of alpha-, it forms beta-glycosidic linkages
While a divalent cation is required for optimal catalytic activity, crystallographic data suggests that the metal and substrate do not occupy the active site at the same time, and therefore, casts doubt on whether the metal is necessary for a non-reaction function, such as product displacement.
Reference Protein and Structure
- Sequence
-
P04547
 (2.4.1.27)
(2.4.1.27)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Enterobacteria phage T4 (Virus)

- PDB
-
1c3j
- T4 PHAGE BETA-GLUCOSYLTRANSFERASE: SUBSTRATE BINDING AND PROPOSED CATALYTIC MECHANISM
(1.88 Å)



- Catalytic CATH Domains
-
3.40.50.2000
 (see all for 1c3j)
(see all for 1c3j)
- Cofactors
- Magnesium(2+) (1)
Enzyme Reaction (EC:2.4.1.27)
Enzyme Mechanism
Introduction
Although previous proposals for a mechanism involve a general acid catalysis requiring two critical residues, a proton donor and a nucleophile/base current models suggest an alternative. BGT binds two substrates, UDP-glucose and 5-HMC. Glucose transfer occurs by a direct nucleophilic attack by the hydroxyl group of 5-HMC to form a beta-glucosidic bond followed by inversion of configuration. A catalytic base, thought to be Asp100 or Glu22, is provided by the enzyme to abstract a proton from the hydroxyl group activating the attack. Multiple contacts are thought to be made by the enzyme on the substrates but as yet are all speculative.
Catalytic Residues Roles
| UniProt | PDB* (1c3j) | ||
| Arg191 | Arg191A | Enzyme activity has been associated with a domain rearrangement, thought to be mediated by the ionic interaction between Asp100 and Arg191. | hydrogen bond donor, electrostatic stabiliser, steric role |
| Phe72 | Phe72A | Phe72 is thought to be important for intercalating into the DNA duplex and anchoring the substrate in place. | steric role, electrostatic stabiliser |
| Asp100, Glu22 | Asp100A, Glu22A | General acid/base. There is some debate in the literature as to whether the base is Asp100 or Glu22. | hydrogen bond acceptor, hydrogen bond donor, proton acceptor, proton donor, activator, electrostatic stabiliser, steric role |
Chemical Components
bimolecular nucleophilic substitution, proton transfer, overall reactant used, overall product formed, inferred reaction step, native state of enzyme regeneratedReferences
- Unligil UM et al. (2000), Curr Opin Struct Biol, 10, 510-517. Glycosyltransferase structure and mechanism. DOI:10.1016/s0959-440x(00)00124-x. PMID:11042447.
- Tvaroška I (2015), Carbohydr Res, 403, 38-47. Atomistic insight into the catalytic mechanism of glycosyltransferases by combined quantum mechanics/molecular mechanics (QM/MM) methods. DOI:10.1016/j.carres.2014.06.017. PMID:25060837.
- Lairson LL et al. (2008), Annu Rev Biochem, 77, 521-555. Glycosyltransferases: structures, functions, and mechanisms. DOI:10.1146/annurev.biochem.76.061005.092322. PMID:18518825.
- Larivière L et al. (2003), J Mol Biol, 330, 1077-1086. Crystal Structures of the T4 Phage β-Glucosyltransferase and the D100A Mutant in Complex with UDP-glucose: Glucose Binding and Identification of the Catalytic Base for a Direct Displacement Mechanism. DOI:10.1016/s0022-2836(03)00635-1. PMID:12860129.
- Moréra S et al. (1999), J Mol Biol, 292, 717-730. T4 Phage β-Glucosyltransferase: Substrate Binding and Proposed Catalytic Mechanism. DOI:10.1006/jmbi.1999.3094. PMID:10497034.
- Murray BW et al. (1997), Biochemistry, 36, 823-831. Mechanism of Human α-1,3-Fucosyltransferase V: Glycosidic Cleavage Occurs Prior to Nucleophilic Attack†. DOI:10.1021/bi962284z. PMID:9020780.
- Davies G et al. (1995), Structure, 3, 853-859. Structures and mechanisms of glycosyl hydrolases. DOI:10.1016/s0969-2126(01)00220-9. PMID:8535779.
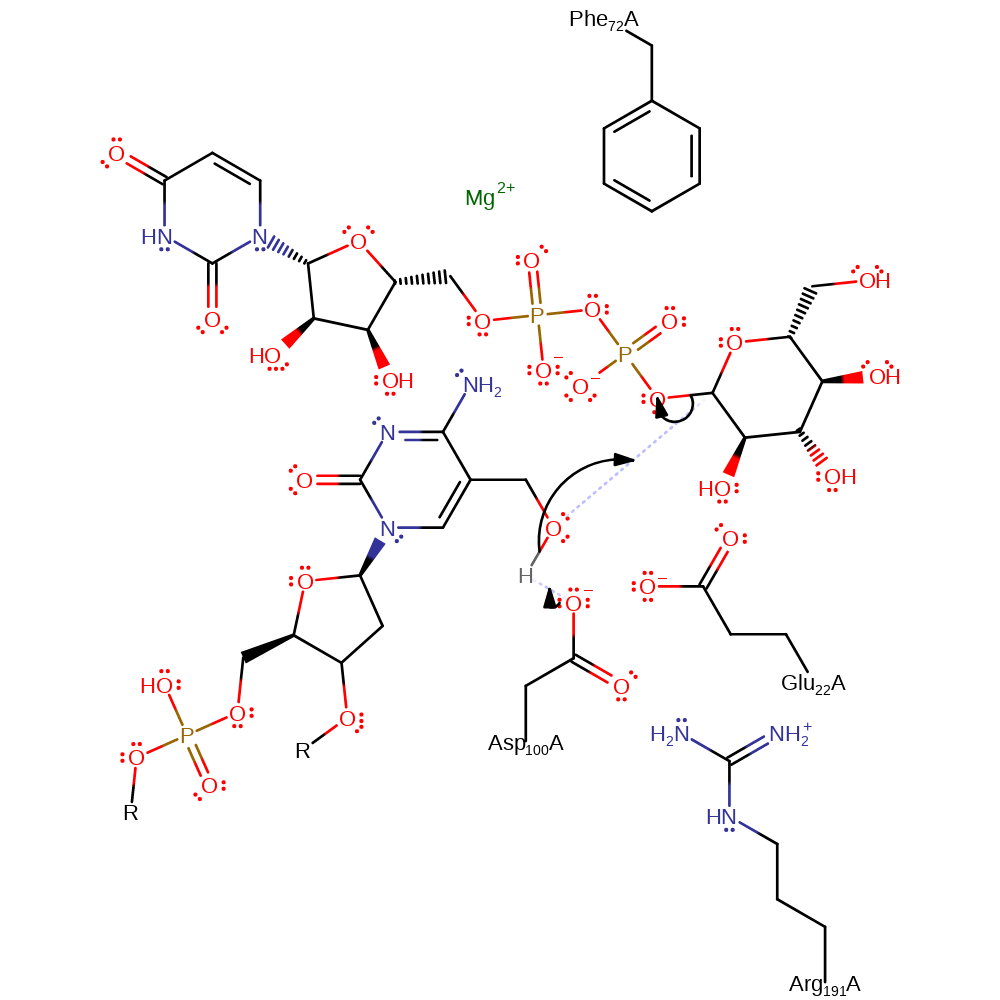
Step 1. BGT binds two substrates, UDP-glucose and 5-HMC. Glucose transfer occurs by a direct nucleophilic attack by the hydroxyl group of 5-HMC to form a beta-glycosidic bond followed by inversion of configuration. A catalytic base, Asp100, is provided by the enzyme to abstract a proton from the hydroxyl group activating the attack. Glucose transfer proceeds via an oxocarbenium ion-character transition state, resulting in an inversion of configuration and the formation of a beta-glycosidic linkage.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp100A | activator, hydrogen bond acceptor, electrostatic stabiliser, steric role |
| Arg191A | hydrogen bond donor, electrostatic stabiliser, steric role |
| Phe72A | steric role, electrostatic stabiliser |
| Glu22A | hydrogen bond acceptor |
| Asp100A | proton acceptor |
Chemical Components
ingold: bimolecular nucleophilic substitution, proton transfer, overall reactant used, overall product formedCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp100A | activator, hydrogen bond donor |
| Arg191A | hydrogen bond donor, electrostatic stabiliser |
| Asp100A | proton donor |






 Download:
Download: