Lactoylglutathione lyase
Lactoyl-glutathione lyase (or glyoxalase I) is part of the glyoxalase system which catalyses the conversion of acyclic alpha-oxoaldehydes into the corresponding alpha-hydroxyacids. Glyoxalase I catalyses the isomerization of the hemithioacetal (formed spontaneously from alpha-oxoaldehyde and GSH), to S-2-hydroxyacylglutathione (or R) derivatives, therefore decreasing the steady-state concentrations of physiological alpha-oxoaldehydes and associated glycation reactions. Physiological substrates of glyoxalase I are methylglyoxal, glyoxal and other acyclic alpha-oxoaldehydes.
This is the first of two steps in the conversion of 2-oxo-aldehydes to the corresponding 2-hydroxycarboxylic acids by way of the glyoxylase system. Methylglyoxal is produced as a by product of the triosephosphate isomerase reaction in glycolysis and, if not removed, is toxic as it reacts readily with with proteins and nucleic acids.
Reference Protein and Structure
- Sequence
-
Q04760
 (4.4.1.5)
(4.4.1.5)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Homo sapiens (Human)

- PDB
-
1qin
- HUMAN GLYOXALASE I COMPLEXED WITH S-(N-HYDROXY-N-P-IODOPHENYLCARBAMOYL) GLUTATHIONE
(2.0 Å)



- Catalytic CATH Domains
-
3.10.180.10
 (see all for 1qin)
(see all for 1qin)
- Cofactors
- Zinc(2+) (1) Metal MACiE
Enzyme Reaction (EC:4.4.1.5)
Enzyme Mechanism
Introduction
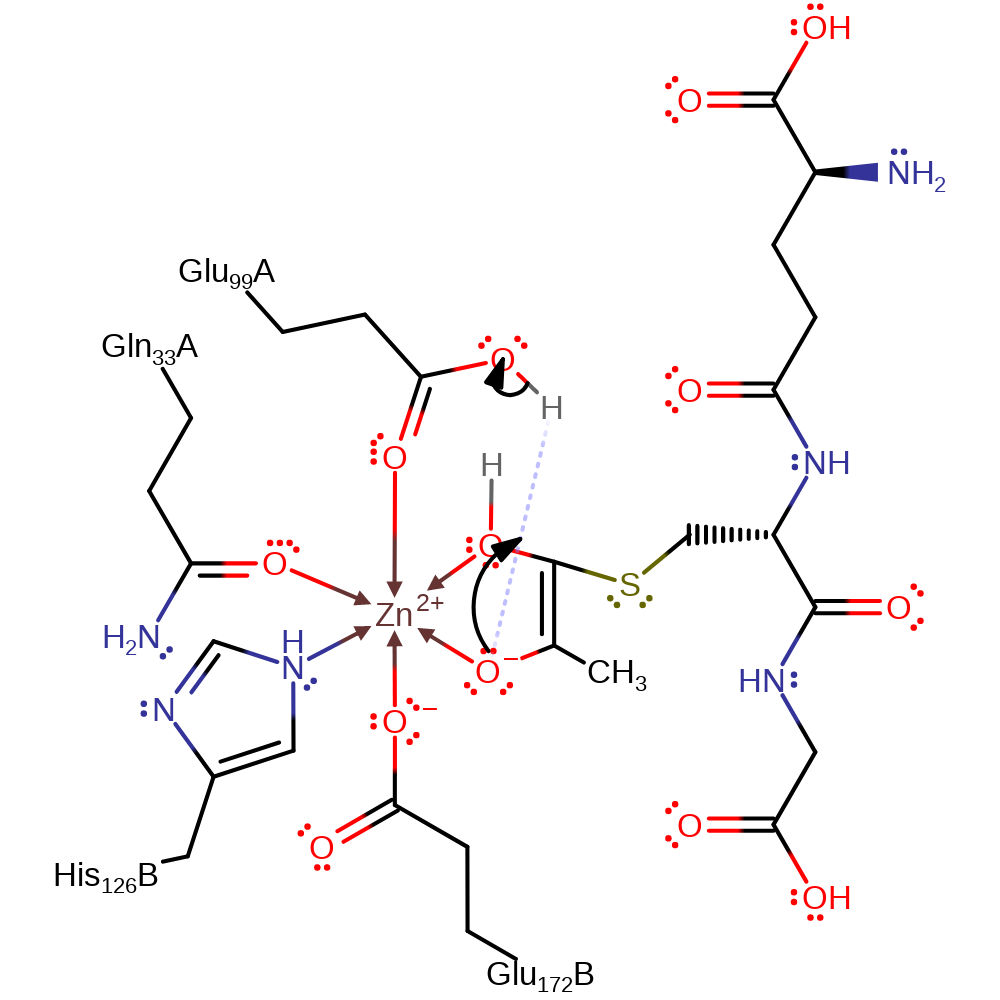
This is the mechanism that occurs with the R enantiomer. The mechanism employed by the enzyme reflects the stereochemistry of the substrate. Both S and R forms of the hemithioacetal bind in the active site, coordinated to the Zn(II) metal in the place of previously coordinated water molecules.
The R enantiomer reaction mechanism differs in that it involves two bases (Glu99 and Glu172) as opposed to only one base in the S case (Glu172). The enzyme only works on the hemithioacetal intermediate which is formed by the spontaneous reaction between methylglyoxal and glutathione.
Both reaction mechanisms form a cis-ene-diol intermediate coordinated directly to the Zn centre. This intermediate then undergoes keto-enol tautomerisation, assisted by Glu172 in both mechanisms to form the R-2-hydroxyacylglutathione product.
Lactoylgutathione lyase I catalyses the first half of the mechanism to detoxify methyl-glyoxyl, where the product of this reaction is relayed to glyoxylase II (M0157) which catalyses the hydrolysis reaction to form D-lactate and glutathione.
Catalytic Residues Roles
| UniProt | PDB* (1qin) | ||
| Glu100 | Glu99A | The residue is correctly positioned to act as a general base towards the R substrate enantiomer. | hydrogen bond acceptor, hydrogen bond donor, metal ligand, proton acceptor, proton donor |
| Glu173 | Glu172B | The residue is positioned correctly to act as a general base towards the S-substrate enantiomer. Glu172 then assists in the keto-enol tautomerisation of both enantiomers in the product forming stage of the reaction. | hydrogen bond acceptor, metal ligand, proton acceptor, proton donor, proton relay |
| His127, Glu100, Glu173, Gln34 | His126B, Glu99A, Glu172B, Gln33A | Binds the Zn(II) ion. | metal ligand |
Chemical Components
reaction occurs outside the enzyme, atom stereo change, cofactor used, assisted keto-enol tautomerisation, overall reactant used, intermediate formation, proton transfer, overall product formed, intermediate terminated, proton relay, native state of enzyme regenerated, native state of cofactor is not regeneratedReferences
- Cameron AD et al. (1999), Biochemistry, 38, 13480-13490. Reaction Mechanism of Glyoxalase I Explored by an X-ray Crystallographic Analysis of the Human Enzyme in Complex with a Transition State Analogue†. DOI:10.1021/bi990696c. PMID:10521255.
- Maroney MJ et al. (2014), Chem Rev, 114, 4206-4228. Nonredox Nickel Enzymes. DOI:10.1021/cr4004488. PMID:24369791.
- Suttisansanee U et al. (2011), Semin Cell Dev Biol, 22, 285-292. Bacterial glyoxalase enzymes. DOI:10.1016/j.semcdb.2011.02.004. PMID:21310258.
- Sukdeo N et al. (2007), Biochim Biophys Acta, 1774, 756-763. Pseudomonas aeruginosa contains multiple glyoxalase I-encoding genes from both metal activation classes. DOI:10.1016/j.bbapap.2007.04.005. PMID:17513180.
- Clugston SL et al. (2004), Biochem J, 377, 309-316. Investigation of metal binding and activation of Escherichia coli glyoxalase I: kinetic, thermodynamic and mutagenesis studies. DOI:10.1042/bj20030271. PMID:14556652.
- Thornalley PJ (2003), Biochem Soc Trans, 31, 1343-1348. Glyoxalase I--structure, function and a critical role in the enzymatic defence against glycation. PMID:14641060.
- Himo F et al. (2001), J Am Chem Soc, 123, 10280-10289. Catalytic Mechanism of Glyoxalase I: A Theoretical Study. DOI:10.1021/ja010715h. PMID:11603978.
- Armstrong RN (2000), Biochemistry, 39, 13625-13632. Mechanistic Diversity in a Metalloenzyme Superfamily†. DOI:10.1021/bi001814v. PMID:11076500.
- Tamaki A et al. (2000), J Nat Prod, 63, 1417-1419. Phenolic Glycosides from the Leaves ofAlangiumplatanifoliumvar.platanifolium. DOI:10.1021/np000119l. PMID:11076566.
- Ridderström M et al. (1998), J Biol Chem, 273, 21623-21628. Involvement of an Active-site Zn2+ Ligand in the Catalytic Mechanism of Human Glyoxalase I. DOI:10.1074/jbc.273.34.21623. PMID:9705294.
- Sellin S et al. (1982), J Biol Chem, 257, 10023-10029. Nuclear relaxation studies of the role of the essential metal in glyoxalase I. PMID:7107595.
Catalytic Residues Roles
| Residue | Roles |
|---|
Chemical Components
reaction occurs outside the enzyme, atom stereo change, cofactor used
Step 2. Glu99 deprotonates the C3 carbon adjacent to the sulfur atom, resulting in double bond rearrangement and formation of the enol-intermediate.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu99A | hydrogen bond acceptor |
| Glu172B | metal ligand, hydrogen bond acceptor |
| Glu99A | metal ligand |
| Gln33A | metal ligand |
| His126B | metal ligand |
| Glu99A | proton acceptor |
Chemical Components
assisted keto-enol tautomerisation, atom stereo change, overall reactant used, intermediate formation, proton transferCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu99A | hydrogen bond donor |
| Glu172B | metal ligand, hydrogen bond acceptor |
| Glu99A | metal ligand |
| Gln33A | metal ligand |
| His126B | metal ligand |
| Glu99A | proton donor |
Chemical Components
intermediate formation, proton transfer
Step 4. Glu172B deprotonates the C3 hydroxy group, causing double bond rearrangement and the protonation of C2 from Glu172B.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu99A | hydrogen bond acceptor |
| Glu172B | hydrogen bond acceptor, proton relay |
| Glu172B | metal ligand |
| Glu99A | metal ligand |
| Gln33A | metal ligand |
| His126B | metal ligand |
| Glu172B | proton acceptor, proton donor |




 Download:
Download: