Riboflavin synthase
Riboflavin (vitamin B2) serves as a precursor of flavocoenzymes, which have essential roles as redox cofactors in all organisms. The final step in the biosynthesis of the vitamin is catalysed by the enzyme riboflavin synthase. This unusual reaction involves the dismutation of 6,7-dimethyl-8-ribityllumazine. A 4-carbon unit is transferred between two of the identical substrates to form riboflavin and 5-amino-6-ribitylamino-2,4(1H,3H)-pyrimidinedione involving the cleavage of two C-N bonds and the formation of two C-C bonds by the transfer of a 4-carbon moiety. The three active sites of the trimer lie between pairs of monomers, although only one active site can be formed and catalytically competent at any one time. The homotrimer is non-existent in humans and is an attractive target for antimicrobial agents.
Reference Protein and Structure
- Sequence
-
Q9Y7P0
 (2.5.1.9)
(2.5.1.9)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Schizosaccharomyces pombe 972h- (Fission yeast)

- PDB
-
1kzl
- Riboflavin Synthase from S.pombe bound to Carboxyethyllumazine
(2.1 Å)



- Catalytic CATH Domains
-
2.40.30.20
 (see all for 1kzl)
(see all for 1kzl)
- Cofactors
- Water (1)
Enzyme Reaction (EC:2.5.1.9)
Enzyme Mechanism
Introduction
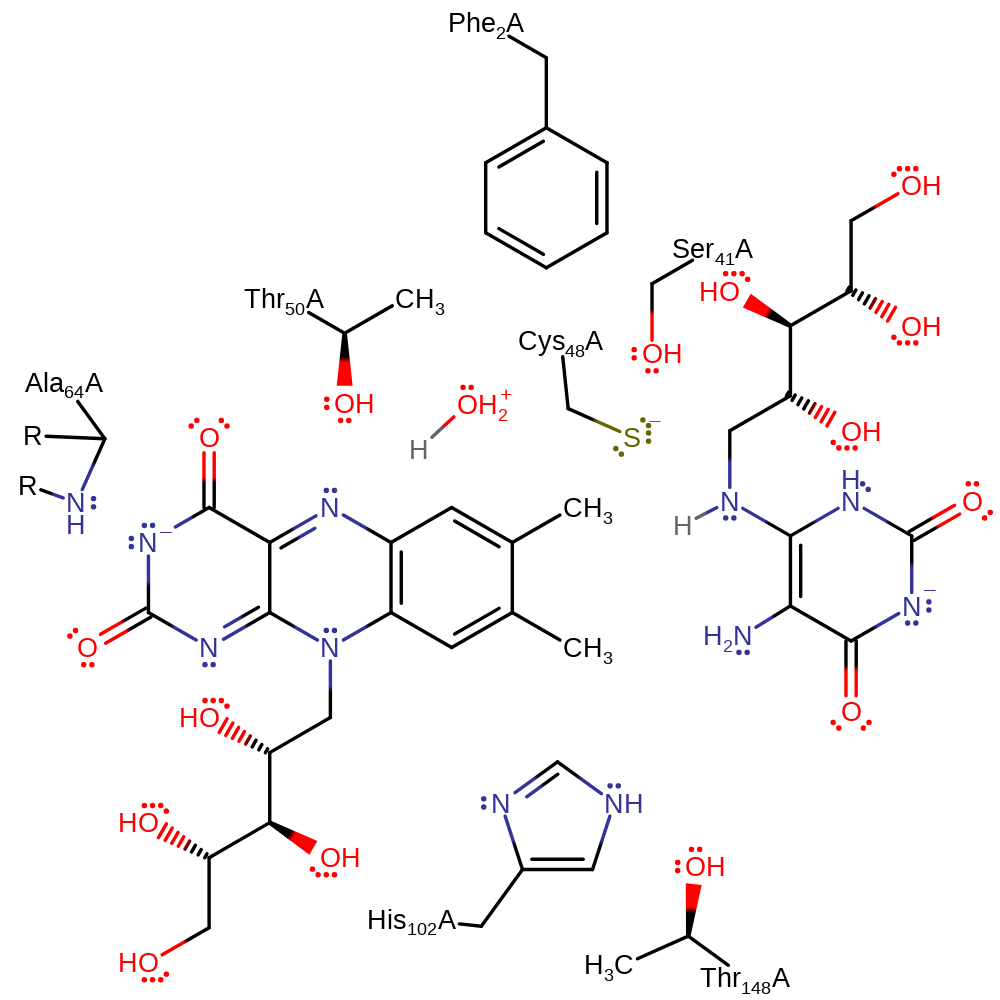
This mechanism is not yet fully characterised, and the roles of the various amino acid residues are still very putative.
In this proposal, His102 deprotonates the 7 position methyl group of the first substrate molecule in an initial activation step. Cys48 initiates a nucleophilic attack on the second substrate in an addition reaction with the concomitant deprotonation of solvent water. (The nucleophile could be a hydroxide formed from water.) The oxyanion of the first activated substrate collapses, initiating a nucleophilic attack on the second activate substrate with concomitant deprotonation of His102. His102 deprotonates the intermediate, initiating a double bond rearrangement, re-forming the oxyanion. The intermediate tautomerises, in two steps. Water deprotonates the amine with concomitant double bond rearrangement which initiates a nucleophilic attack on the carbon to which the Cys48 is covalently attached in a substitution reaction. The oxyanion collapses, initiating double bond rearrangement that results in the cleavage of the C-N bond in an intramolecular elimination with concomitant deprotonation of His102. Cys48 deprotonates the ring CH2 group, initiating an elimination reaction, which liberates the reaction products with concomitant deprotonation of Cys48.
The role of individual amino acids is still putative.
Catalytic Residues Roles
| UniProt | PDB* (1kzl) | ||
| Phe2 | Phe2A | The Phe2 side chain stabilises the Cys48 thiol so that nucleophilic attack can occur. | increase nucleophilicity, enhance reactivity, electrostatic stabiliser, increase acidity |
| Ser41 | Ser41A | Aids E2 elimination by Cys48, could potentially act as a general acid. Ser41 also stabilises the Cys48 thiolate ion forming a hydrogen bond to the cysteine residue. | hydrogen bond donor, electrostatic stabiliser, increase acidity, increase basicity, increase nucleophilicity |
| Cys48 | Cys48A | Cys48 thiol group nucleophilically attacks the 7-position of one of the lumazine substrate molecules, and is stabilised by hydrogen bonding to Ser 41 and Phe 2. Also acts as a general acid/base in the final steps of the mechanism in the E2 elimination that forms the products. | covalently attached, hydrogen bond acceptor, hydrogen bond donor, nucleophile, proton acceptor, proton donor, nucleofuge, activator, increase electrophilicity |
| Thr50 | Thr50A | Thought to activate the second tautomerisation. | increase basicity, hydrogen bond donor, electrostatic stabiliser, increase acidity |
| His102 | His102A | Acts in conjunction with Thr148 as a dyad in general base/acid function. Initiates formation of oxyanion via deprotonation of DMRL. Initiates tautomerisation. | hydrogen bond acceptor, hydrogen bond donor, proton acceptor, proton donor |
| Thr148 | Thr148A | Aids His102 in its function as a general acid/base. | increase basicity, hydrogen bond acceptor, electrostatic stabiliser, increase acidity |
| Ala64 (main-N) | Ala64A (main-N) | Stabilises the exo-methylene anion by hydrogen bonding via its backbone amide. The identity of this residue doesn't seem to be conserved (Met in the E. coli protein, Ala in the S. pombe protein). | hydrogen bond donor, electrostatic stabiliser |
Chemical Components
proton transfer, overall reactant used, intermediate formation, bimolecular nucleophilic addition, enzyme-substrate complex formation, intramolecular rearrangement, tautomerisation (not keto-enol), intermediate terminated, intramolecular nucleophilic substitution, enzyme-substrate complex cleavage, cyclisation, intramolecular elimination, decyclisation, bimolecular elimination, overall product formed, native state of enzyme regeneratedReferences
- Fischer M et al. (2008), Arch Biochem Biophys, 474, 252-265. Biosynthesis of vitamin B2: Structure and mechanism of riboflavin synthase. DOI:10.1016/j.abb.2008.02.008. PMID:18298940.
- Kim RR et al. (2010), J Am Chem Soc, 132, 2983-2990. Mechanistic Insights on Riboflavin Synthase Inspired by Selective Binding of the 6,7-Dimethyl-8-ribityllumazine Exomethylene Anion. DOI:10.1021/ja908395r. PMID:20143812.
- Fischer M et al. (2005), Nat Prod Rep, 22, 324-350. Biosynthesis of flavocoenzymes. DOI:10.1039/b210142b. PMID:16010344.
- Illarionov B et al. (2005), Biol Chem, 386, 127-136. Pre-steady-state kinetic analysis of riboflavin synthase using a pentacyclic reaction intermediate as substrate. DOI:10.1515/bc.2005.016. PMID:15843156.
- Zheng YJ et al. (2003), Bioorg Chem, 31, 278-287. Examination of a reaction intermediate in the active site of riboflavin synthase. DOI:10.1016/s0045-2068(03)00029-4. PMID:12877878.
- Fischer M et al. (2003), BMC Biochem, 4, 18-. Riboflavin synthase of Schizosaccharomyces pombe. Protein dynamics revealed by 19F NMR protein perturbation experiments. DOI:10.1186/1471-2091-4-18. PMID:14690539.
- Liao DI et al. (2001), Structure, 9, 399-408. Crystal Structure of Riboflavin Synthase. DOI:10.1016/s0969-2126(01)00600-1. PMID:11377200.
- Truffault V et al. (2001), J Mol Biol, 309, 949-960. The solution structure of the N-terminal domain of riboflavin synthase. DOI:10.1006/jmbi.2001.4683. PMID:11399071.
- Illarionov B et al. (2001), J Biol Chem, 276, 11524-11530. Riboflavin Synthase of Escherichia coli. EFFECT OF SINGLE AMINO ACID SUBSTITUTIONS ON REACTION RATE AND LIGAND BINDING PROPERTIES. DOI:10.1074/jbc.m008931200. PMID:11278450.
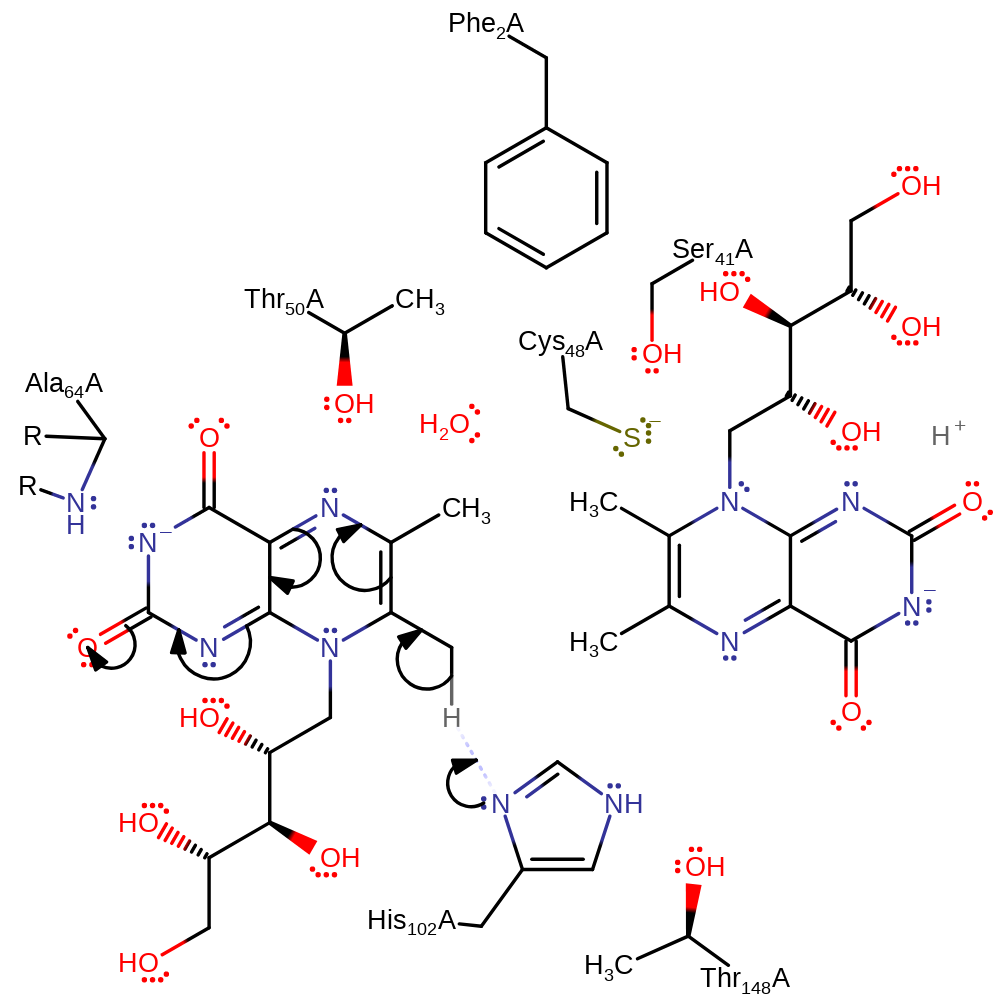
Step 1. His102 deprotonates the 7 position methyl group of the first substrate molecule in an initial activation step.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Thr148A | hydrogen bond acceptor, increase basicity |
| Ser41A | hydrogen bond donor, electrostatic stabiliser |
| His102A | hydrogen bond donor, hydrogen bond acceptor |
| Cys48A | hydrogen bond acceptor |
| Thr50A | hydrogen bond donor |
| Ala64A (main-N) | hydrogen bond donor |
| Phe2A | electrostatic stabiliser |
| His102A | proton acceptor |
Chemical Components
proton transfer, overall reactant used, intermediate formation
Step 2. Cys48 initiates a nucleophilic attack on the second substrate in an addition reaction with the concomitant deprotonation of solvent water.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Thr148A | hydrogen bond acceptor |
| Ser41A | increase nucleophilicity, hydrogen bond donor |
| His102A | hydrogen bond donor |
| Cys48A | hydrogen bond acceptor |
| Thr50A | hydrogen bond donor |
| Phe2A | increase nucleophilicity |
| Ala64A (main-N) | electrostatic stabiliser |
| Cys48A | nucleophile |
Chemical Components
ingold: bimolecular nucleophilic addition, proton transfer, overall reactant used, intermediate formation, enzyme-substrate complex formation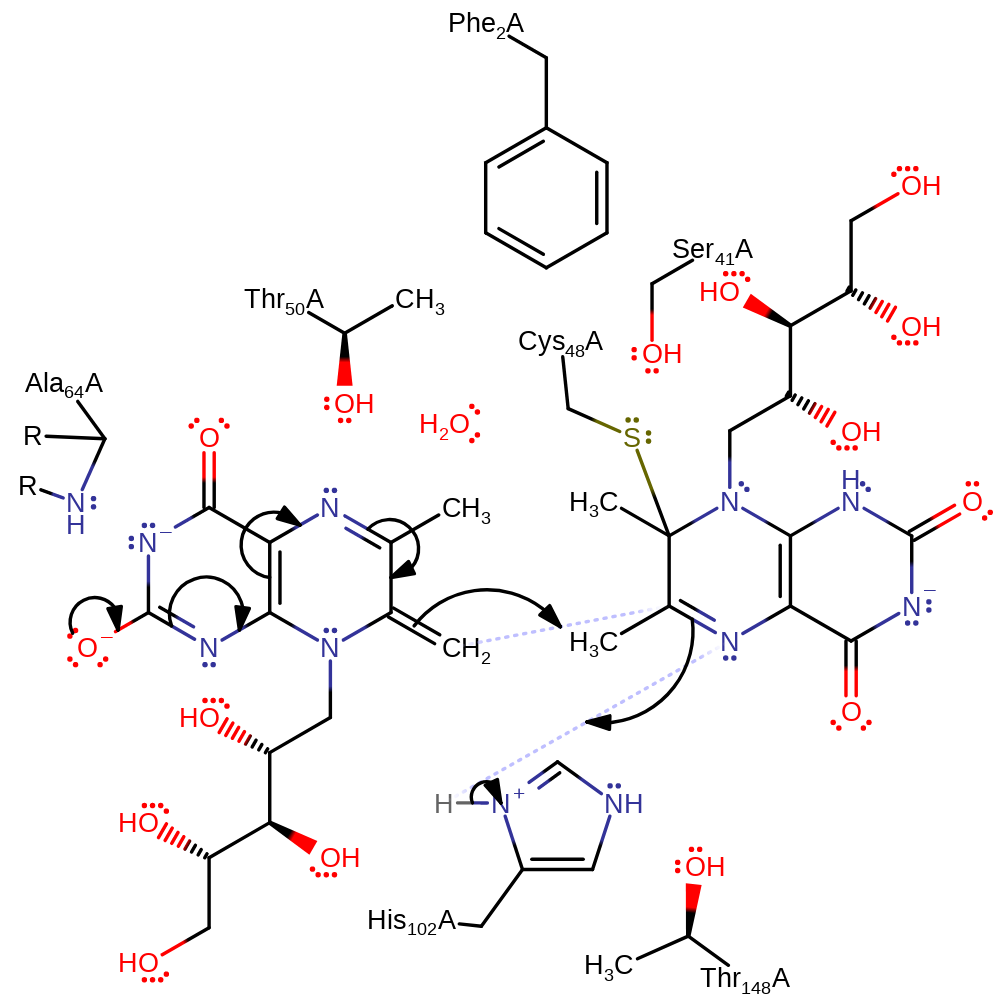
Step 3. The oxyanion of the first activated substrate collapses, initiating a nucleophilic attack on the second activate substrate with concomitant deprotonation of His102.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Thr148A | increase acidity, hydrogen bond acceptor |
| His102A | hydrogen bond donor |
| Cys48A | covalently attached, increase electrophilicity |
| Thr50A | hydrogen bond donor |
| Ala64A (main-N) | electrostatic stabiliser |
| His102A | proton donor |
Chemical Components
ingold: bimolecular nucleophilic addition, proton transfer, intermediate formation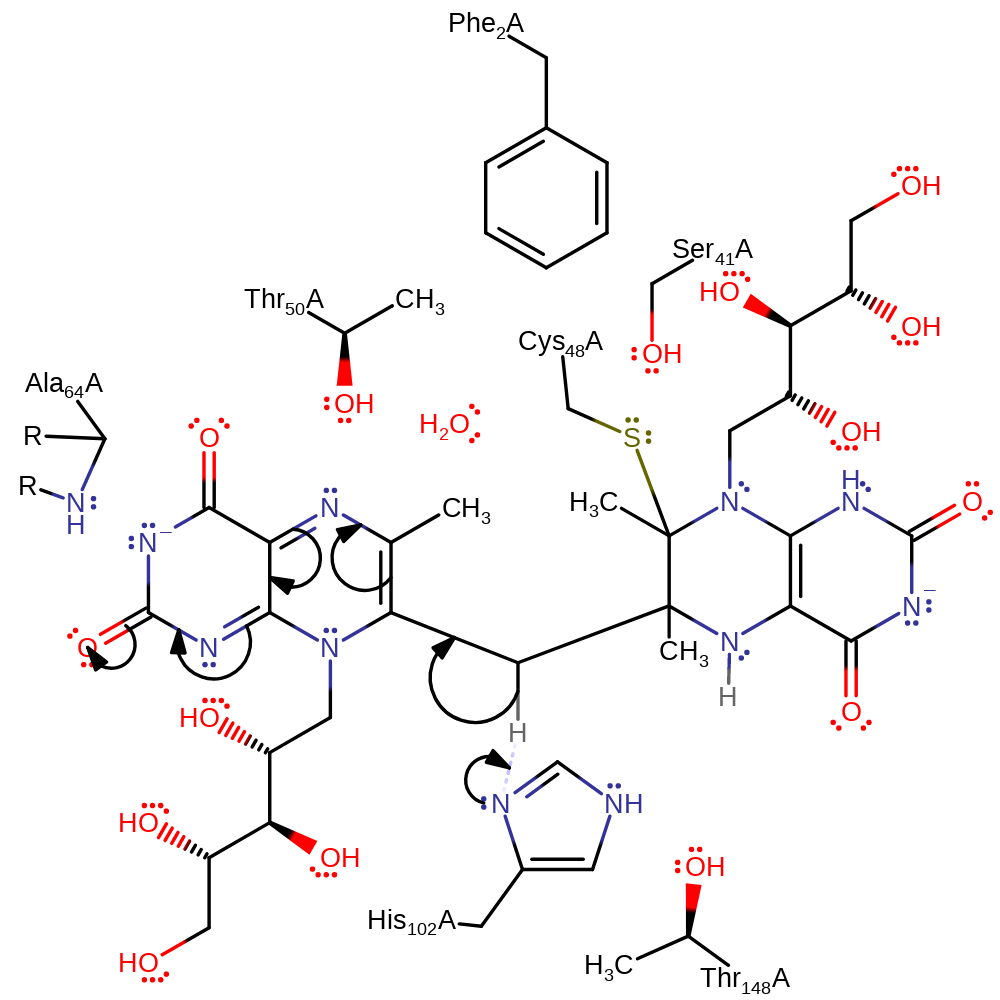
Step 4. His102 deprotonates the intermediate, initiating a double bond rearrangement, re-forming the oxyanion.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Thr148A | increase basicity |
| His102A | hydrogen bond acceptor, hydrogen bond donor |
| Cys48A | covalently attached, activator |
| Thr50A | hydrogen bond donor |
| Ala64A (main-N) | electrostatic stabiliser |
| His102A | proton acceptor |
Chemical Components
proton transfer, intramolecular rearrangement, intermediate formationCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Thr148A | hydrogen bond acceptor |
| His102A | hydrogen bond donor |
| Cys48A | covalently attached, activator |
| Thr50A | increase basicity, hydrogen bond donor |
Chemical Components
tautomerisation (not keto-enol), intermediate terminated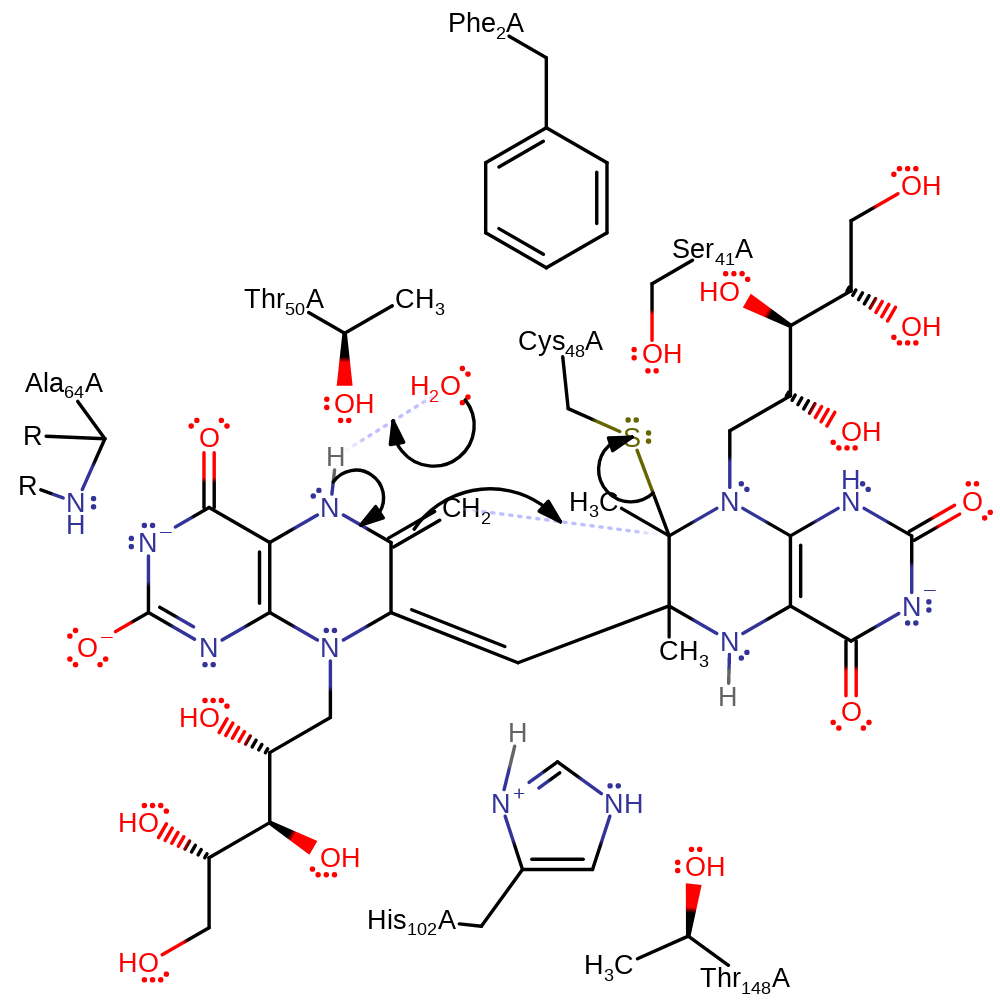
Step 6. Water deprotonates the amine with concomitant double bond rearrangement which initiates a nucleophilic attack on the carbon to which the Cys48 is covalently attached in a substitution reaction.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Thr148A | hydrogen bond acceptor |
| Ser41A | hydrogen bond donor, electrostatic stabiliser |
| His102A | hydrogen bond donor |
| Cys48A | covalently attached, hydrogen bond acceptor |
| Thr50A | increase acidity, hydrogen bond donor |
| Ala64A (main-N) | electrostatic stabiliser |
| Cys48A | nucleofuge |
Chemical Components
proton transfer, ingold: intramolecular nucleophilic substitution, enzyme-substrate complex cleavage, intermediate formation, cyclisation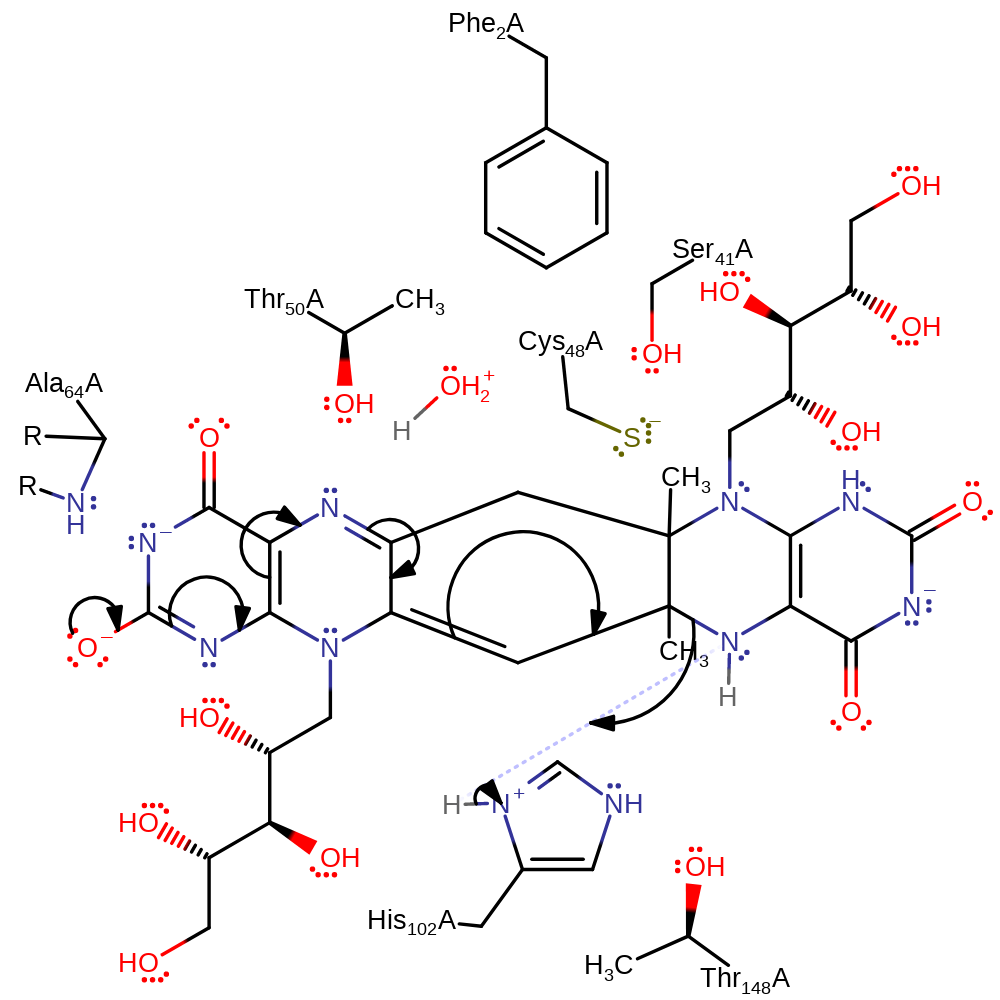
Step 7. The oxyanion collapses, initiating double bond rearrangement that results in the cleavage of the C-N bond in an intramolecular elimination with concomitant deprotonation of His102.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Thr148A | increase basicity, hydrogen bond acceptor |
| Ser41A | hydrogen bond donor |
| His102A | hydrogen bond donor |
| Cys48A | hydrogen bond acceptor |
| Thr50A | hydrogen bond donor |
| Ala64A (main-N) | electrostatic stabiliser |
| Phe2A | electrostatic stabiliser |
| His102A | proton donor |
Chemical Components
proton transfer, intramolecular rearrangement, ingold: intramolecular elimination, decyclisation, intermediate formation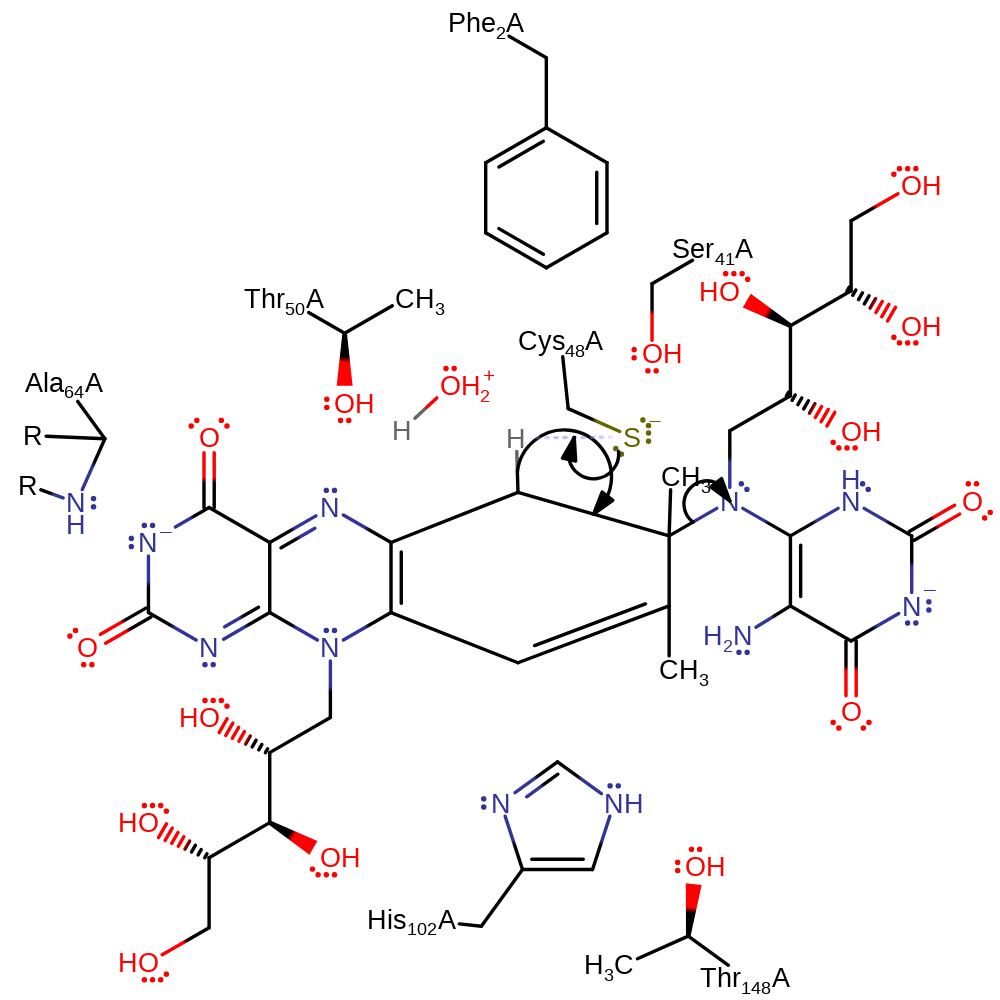
Step 8. Cys48 deprotonates the ring CH2 group, initiating an elimination reaction, which liberates the reaction products.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Thr148A | hydrogen bond acceptor |
| Ser41A | hydrogen bond donor, increase basicity |
| His102A | hydrogen bond donor |
| Cys48A | hydrogen bond acceptor, hydrogen bond donor |
| Thr50A | hydrogen bond donor |
| Phe2A | enhance reactivity |
| Ala64A (main-N) | electrostatic stabiliser |
| Cys48A | proton acceptor |
Chemical Components
ingold: bimolecular elimination, overall product formed, proton transfer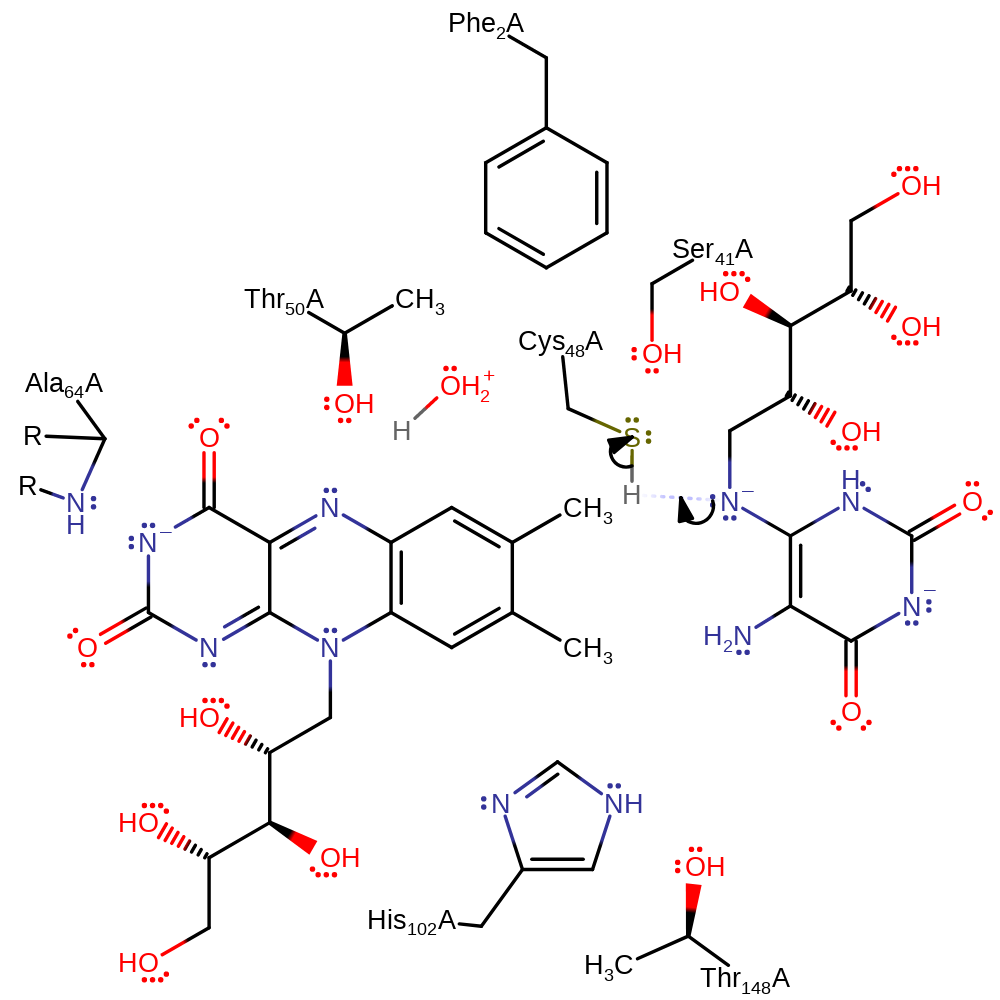
Step 9. This step probably occurs in a concerted manner with step 8 (shown separately for clarity). The riboflavin product deprotonates Cys48 to regenerate the active site and produce the final product.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Phe2A | increase acidity |
| Ser41A | increase acidity |
| Thr50A | electrostatic stabiliser |
| Ala64A (main-N) | electrostatic stabiliser |
| Thr148A | electrostatic stabiliser |
| Cys48A | proton donor |




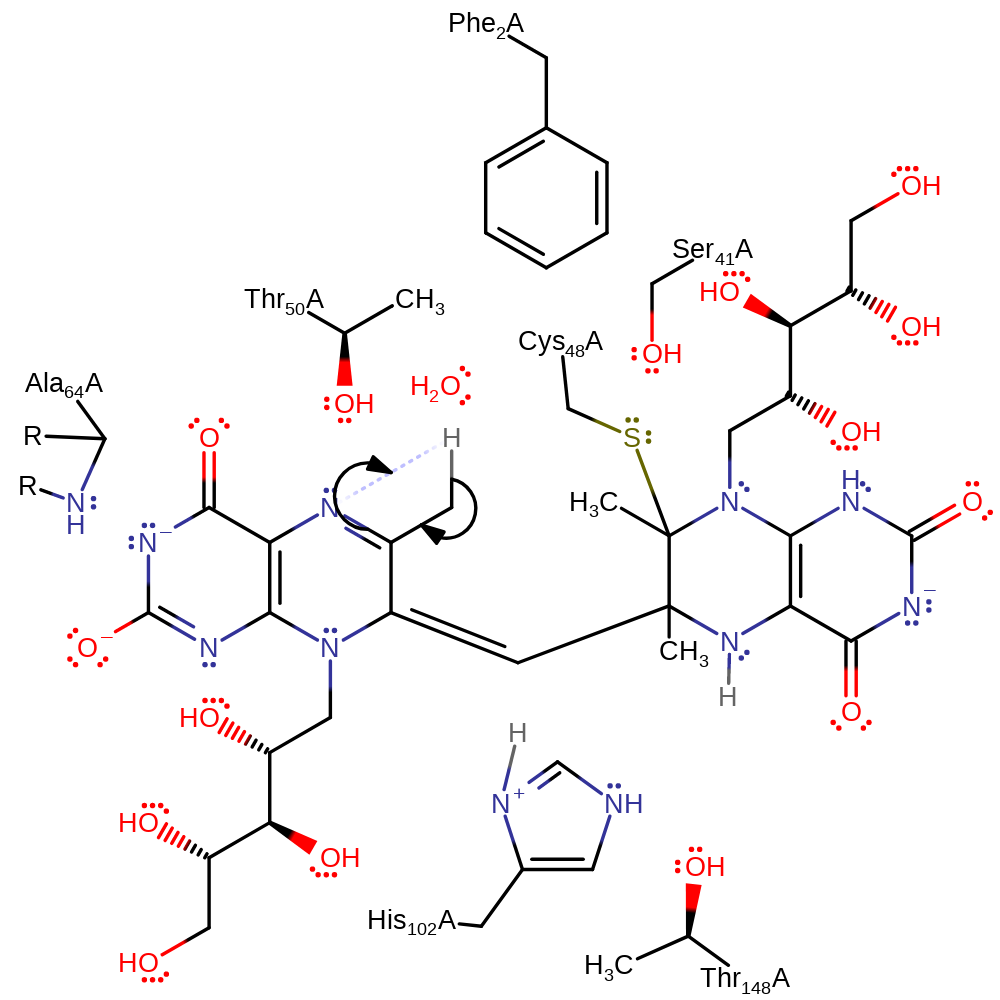
 Download:
Download: