Histone/protein deacetylase
hST2, isolated from Saccharomyces cerevisiae, is a class III histone deacetylase (HDAC), though it also deacetylates other proteins. It belongs to the sirtuin (Sir2) family and catalyses the deacetylation of lysine residues from proteins using NAD+ as a co-substrate. The reaction produces a deacetylated lysine, nicotinamide and 2'-O-acetyl ADP-ribose. hST2 play roles in gene silencing and mediating life span extension. hST2 is of interest because its homologues in humans are potential drug targets for diseases associated with ageing, such as type-II diabetes, and Alzheimer's and Parkinson's diseases.
Reference Protein and Structure
- Sequence
-
P53686
 (2.3.1.286)
(2.3.1.286)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Saccharomyces cerevisiae S288c (Baker's yeast)

- PDB
-
1szd
- Structural basis for nicotinamide cleavage and ADP-ribose transfer by NAD+-dependent Sir2 histone/protein deacetylases
(1.5 Å)



- Catalytic CATH Domains
-
3.30.1600.10
 3.40.50.1220
3.40.50.1220  (see all for 1szd)
(see all for 1szd)
Enzyme Mechanism
Introduction
Aided by lone pair donation of the acetylated lysine nitrogen, the acetyl oxygen acts as the nucleophile for associative, Sn2 attack on the C1' position of the nicotinamide-ribose of NAD+. The leaving group is nicotinamide and the reaction produces a 1'-O-alkylamidate intermediate. His135 deprotonates the 3'-hydroxyl, which then goes on to deprotonate the 2'-hydroxyl. The 2'-hydroxyl is then the nucleophile for attack on the acetyl carbon, forming a 1',2'-cyclic intermediate. The nitrogen of the lysine is protonated by His135 and then dissociates from the intermediate to form a cyclic acyl-oxonium ion. Water attacks the positively-charged carbon (stabilised by the two oxygens of the intermediate) with the lysine of the product deprotonating the positively-charged oxygen. The ring is opened when the exocyclic oxygen forms a carbonyl, with proton transfer to the 1' endocyclic oxygen. This produces 2'-O-acetyl ADP-ribose.
Catalytic Residues Roles
| UniProt | PDB* (1szd) | ||
| His135 | His135(138)A | His135 deprotonates the 3'-hydroxyl of the 1'-O-alkylamidate intermediate. It protonates the nitrogen of the lysine in the 1',2'-cyclic intermediate, making it a better leaving group. | hydrogen bond acceptor, hydrogen bond donor, proton acceptor, proton donor |
| Asp118 | Asp118(121)A | Asp118 forms a hydrogen bond to the carboxamide nitrogen of NAD+. This interaction causes the carboxamide to rotate by 150 degrees and so disrupts the electronic resonance between the carboxamide and the pyridine ring. This weakens the glycosidic bond and destabilises the ground state. Asp118 also interacts with the the 2'-hydroxyl group of the ribose, which may aid NAD+ cleavage by inductive polarisation. | activator, hydrogen bond acceptor |
| Asp43 (main-N) | Asp43(46)A (main-N) | Asp43 is part of a flexible loop that occludes the nicotinamide binding pocket after NAD+ cleavage. This prevents nicotinamide exchange. Asp43 also forms a hydrogen bond to the nicotinamide nitrogen after NAD+ cleavage. This holds the nicotinamide in a non-reactive position, thus preventing nicotinamide exchange. | hydrogen bond acceptor, hydrogen bond donor, steric role |
| Pro42 (main-C) | Pro42(45)A (main-C) | Pro42 is part of a flexible loop that occludes the nicotinamide binding pocket after NAD+ cleavage. This prevents nicotinamide exchange. Pro42 also forms a hydrogen bond to the nicotinamide nitrogen after NAD+ cleavage. This holds the nicotinamide in a non-reactive position, thus preventing nicotinamide exchange. | hydrogen bond acceptor, steric role |
| Arg45 | Arg45(48)A | Arg45 is thought to interact with the O4' of ribose in the 1'-O-alkylamidate intermediate. This is thought to stabilise the high-energy intermediate. | hydrogen bond donor, electrostatic stabiliser |
| Phe44 (main-N), Phe44 | Phe44(47)A (main-N), Phe44(47)A | Phe44 protects the substrate during the formation of the 1'-O-alkylamidate intermediate, preventing hydrolysis. It may also play a part in stabilising the transition state of this reaction. Phe44 can then move between the intermediate and nicotinamide, preventing the nicotinamide exchange reaction. It also forms a hydrogen bond to the nitrogen of nicotinamide, holding it in the non-reactive conformation and preventing nicotinamide exchange. | van der waals interaction, hydrogen bond donor |
| Asn116 | Asn116(119)A | Asn116 forms a water-mediated hydrogen bond to the ribose oxygen of NAD+. This may stabilise the transition state of NAD+ cleavage. The residue also interacts with the the 2'-hydroxyl group of the ribose, which may aid NAD+ cleavage by inductive polarisation. Asn116 also positions, and possibly activates, a water molecule for attack on the cyclic acyl-oxonium intermediate. | hydrogen bond acceptor, hydrogen bond donor, activator, electrostatic stabiliser, increase nucleophilicity |
Chemical Components
bimolecular nucleophilic substitution, intermediate formation, overall product formed, overall reactant used, proton transfer, intramolecular nucleophilic addition, cyclisation, proton relay, heterolysis, charge delocalisation, bimolecular nucleophilic addition, unimolecular elimination by the conjugate base, decyclisation, native state of enzyme regenerated, intermediate terminatedReferences
- Smith BC et al. (2006), Biochemistry, 45, 272-282. Sir2 Protein Deacetylases: Evidence for Chemical Intermediates and Functions of a Conserved Histidine†. DOI:10.1021/bi052014t. PMID:16388603.
- Shi Y et al. (2013), J Phys Chem Lett, 4, 491-495. Sirtuin Deacetylation Mechanism and Catalytic Role of the Dynamic Cofactor Binding Loop. DOI:10.1021/jz302015s. PMID:23585919.
- Sauve AA et al. (2012), Curr Opin Chem Biol, 16, 535-543. Sirtuins: NAD(+)-dependent deacetylase mechanism and regulation. DOI:10.1016/j.cbpa.2012.10.003. PMID:23102634.
- Smith BC et al. (2007), J Am Chem Soc, 129, 5802-5803. Sir2 Deacetylases Exhibit Nucleophilic Participation of Acetyl-Lysine in NAD+Cleavage. DOI:10.1021/ja070162w. PMID:17439123.
- Smith BC et al. (2007), J Biol Chem, 282, 37256-37265. Acetyl-lysine Analog Peptides as Mechanistic Probes of Protein Deacetylases. DOI:10.1074/jbc.m707878200. PMID:17951578.
- Hoff KG et al. (2006), Structure, 14, 1231-1240. Insights into the Sirtuin Mechanism from Ternary Complexes Containing NAD+ and Acetylated Peptide. DOI:10.1016/j.str.2006.06.006. PMID:16905097.
- Khan AN et al. (2006), J Biol Chem, 281, 11702-11711. Use of Substrate Analogs and Mutagenesis to Study Substrate Binding and Catalysis in the Sir2 Family of NAD-dependent Protein Deacetylases. DOI:10.1074/jbc.m511482200. PMID:16520376.
- Avalos JL et al. (2005), Mol Cell, 17, 855-868. Mechanism of Sirtuin Inhibition by Nicotinamide: Altering the NAD+ Cosubstrate Specificity of a Sir2 Enzyme. DOI:10.1016/j.molcel.2005.02.022. PMID:15780941.
- Avalos JL et al. (2004), Mol Cell, 13, 639-648. Structural basis for the mechanism and regulation of Sir2 enzymes. DOI:10.2210/pdb1s7g/pdb. PMID:15023335.
- Zhao K et al. (2004), Proc Natl Acad Sci U S A, 101, 8563-8568. Structural basis for nicotinamide cleavage and ADP-ribose transfer by NAD+-dependent Sir2 histone/protein deacetylases. DOI:10.1073/pnas.0401057101. PMID:15150415.
- Zhao K et al. (2003), Structure, 11, 1403-1411. Structure of the yeast Hst2 protein deacetylase in ternary complex with 2'-O-acetyl ADP ribose and histone peptide. DOI:10.2210/pdb1q1a/pdb. PMID:14604530.
- Min J et al. (2001), Cell, 105, 269-279. Crystal structure of a SIR2 homolog-NAD complex. DOI:10.2210/pdb1ici/pdb. PMID:11336676.
- Finnin MS et al. (2001), Nat Struct Biol, 8, 621-625. Structure of the histone deacetylase SIRT2. DOI:10.1038/89668. PMID:11427894.
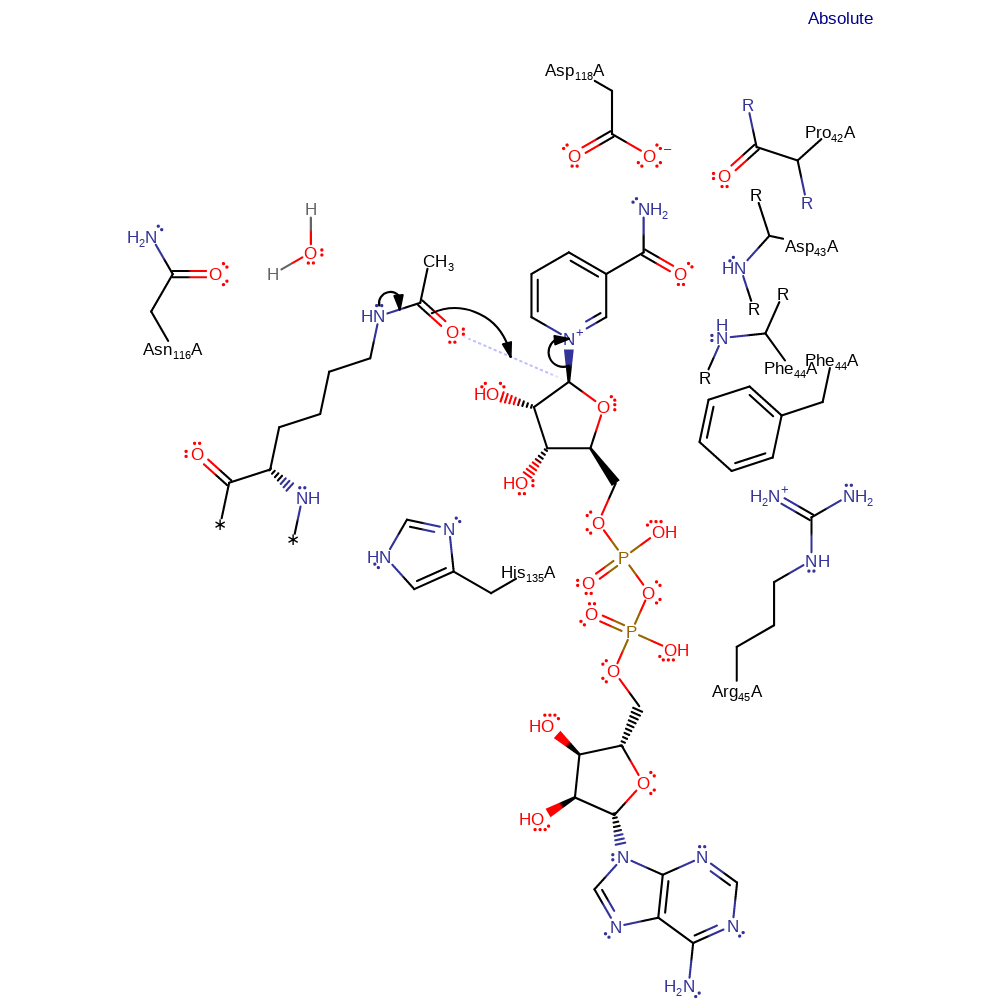
Step 1. With the aid of nitrogen lone pair donation, the acetyl oxygen of acetyllysine is the nucleophile for nucleophilic attack on the C1' position of NAD+, resulting in a substitution reaction in which nicotinamide is the leaving group. This produces a 1'-O-alkylamidate intermediate. The ground state of NAD+ is destabilised because the positively-charged nicotinamide group is positioned in a hydrophobic pocket. Also, Asp118 forces the carboxamide of NAD+ to rotate 150 degrees from its lower energy conformer and this disrupts electronic resonance between the carboxamide and the pyridine ring. After NAD+ cleavage Phe44 moves between the two products to prevent nicotinamide exchange. Nicotinamide rotates around to carboxamide group to form interactions between the nitrogen of the ring and residues. This prevents nicotinamide exchange by positioning the nitrogen nucleophile away from the the intermediate.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Pro42(45)A (main-C) | steric role, hydrogen bond acceptor |
| Arg45(48)A | hydrogen bond donor, electrostatic stabiliser |
| Asn116(119)A | hydrogen bond acceptor, hydrogen bond donor, activator, electrostatic stabiliser |
| His135(138)A | hydrogen bond acceptor |
| Asp118(121)A | hydrogen bond acceptor, activator |
| Phe44(47)A (main-N) | hydrogen bond donor |
| Asp43(46)A (main-N) | steric role, hydrogen bond donor |
| Phe44(47)A | steric role, van der waals interaction |
Chemical Components
ingold: bimolecular nucleophilic substitution, intermediate formation, overall product formed, overall reactant used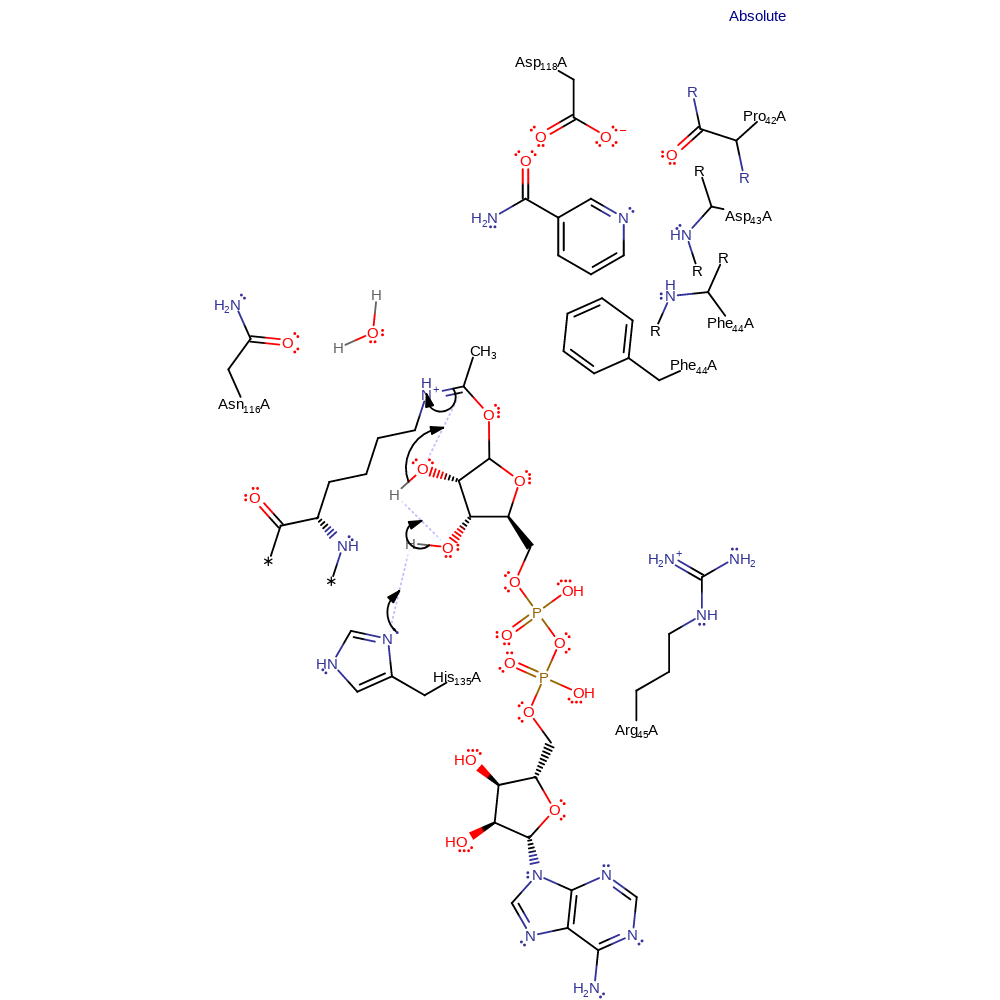
Step 2. His135 deprotonates the 3'-hydroxyl of the 1'-O-alkylamidate intermediate, which then itself deprotonates the 2'-hydroxyl. The resulting alkoxide is the nucleophile for intramolecular nucleophilic attack on the acetyl carbon. This produces a 1',2'-cyclic intermediate.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Phe44(47)A (main-N) | hydrogen bond donor |
| Asp43(46)A (main-N) | hydrogen bond acceptor, steric role |
| Arg45(48)A | hydrogen bond donor |
| Asn116(119)A | hydrogen bond acceptor |
| His135(138)A | hydrogen bond acceptor |
| Pro42(45)A (main-C) | hydrogen bond acceptor, steric role |
| Asp118(121)A | hydrogen bond acceptor |
| Phe44(47)A | steric role, van der waals interaction |
| His135(138)A | proton acceptor |
Chemical Components
proton transfer, ingold: intramolecular nucleophilic addition, cyclisation, intermediate formation, proton relay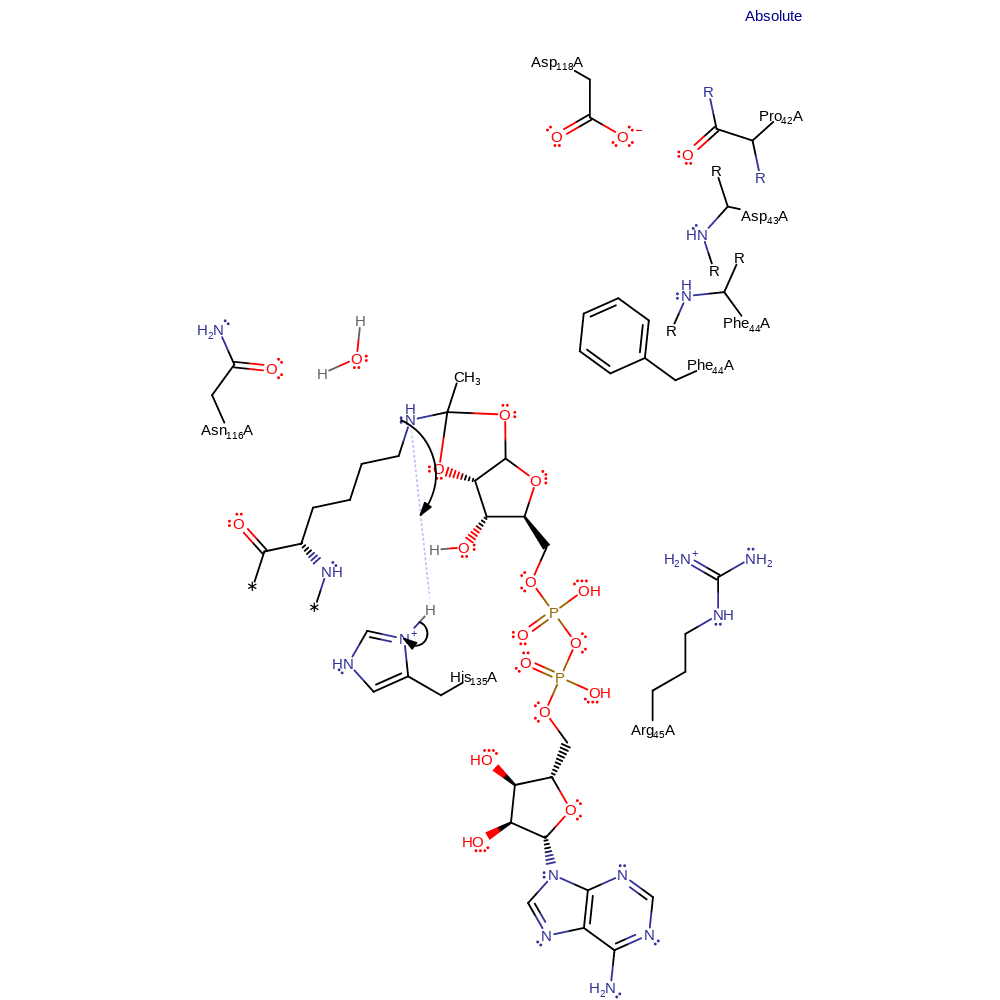
Step 3. Nicotinamide can dissociate from the enzyme after NAD+ cleavage. It has been shown to leave before this step so that the previous step can illustrate how the enzyme prevents nicotinamide exchange. His135 protonates the lysine nitrogen in the 1',2'-cyclic intermediate.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Phe44(47)A (main-N) | van der waals interaction, hydrogen bond donor |
| Arg45(48)A | hydrogen bond donor |
| His135(138)A | hydrogen bond donor, proton donor |
Chemical Components
proton transfer, intermediate formationCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Phe44(47)A (main-N) | van der waals interaction, hydrogen bond donor |
| Arg45(48)A | hydrogen bond donor |
| His135(138)A | hydrogen bond acceptor |
Chemical Components
heterolysis, charge delocalisation, intermediate formation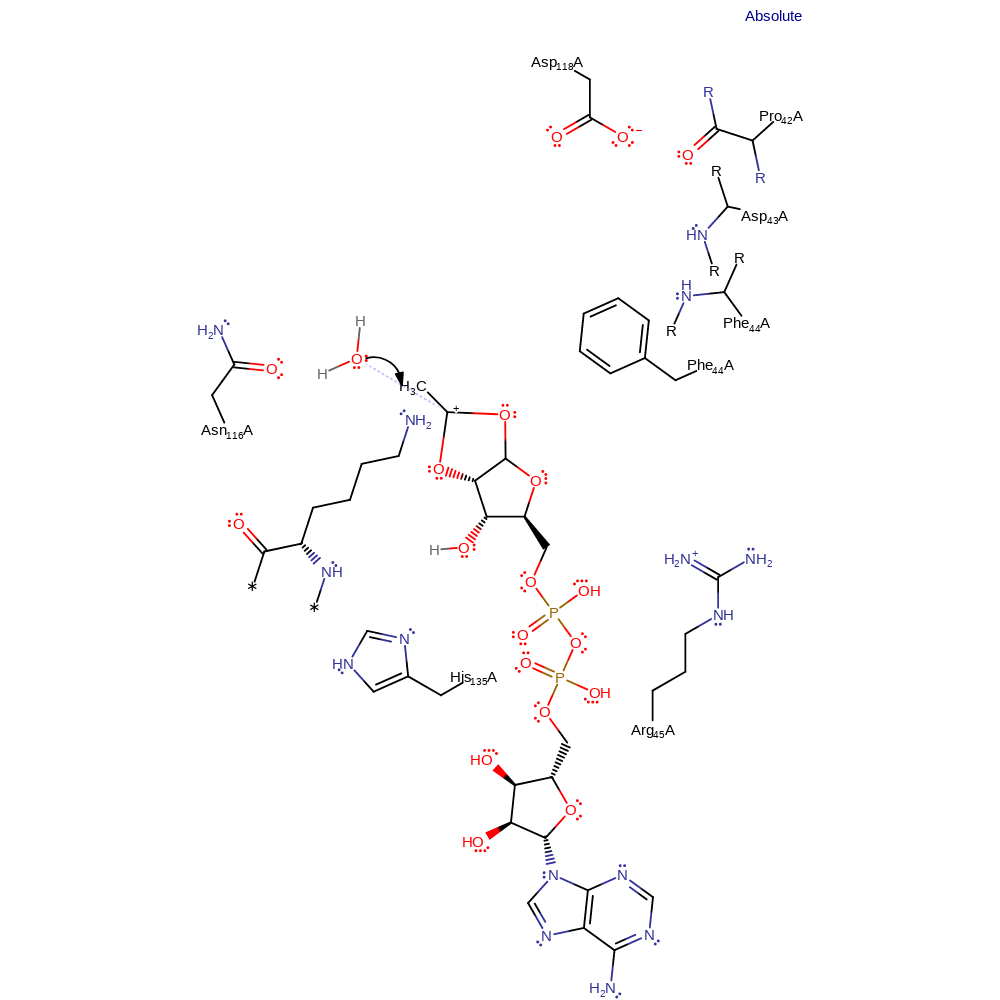
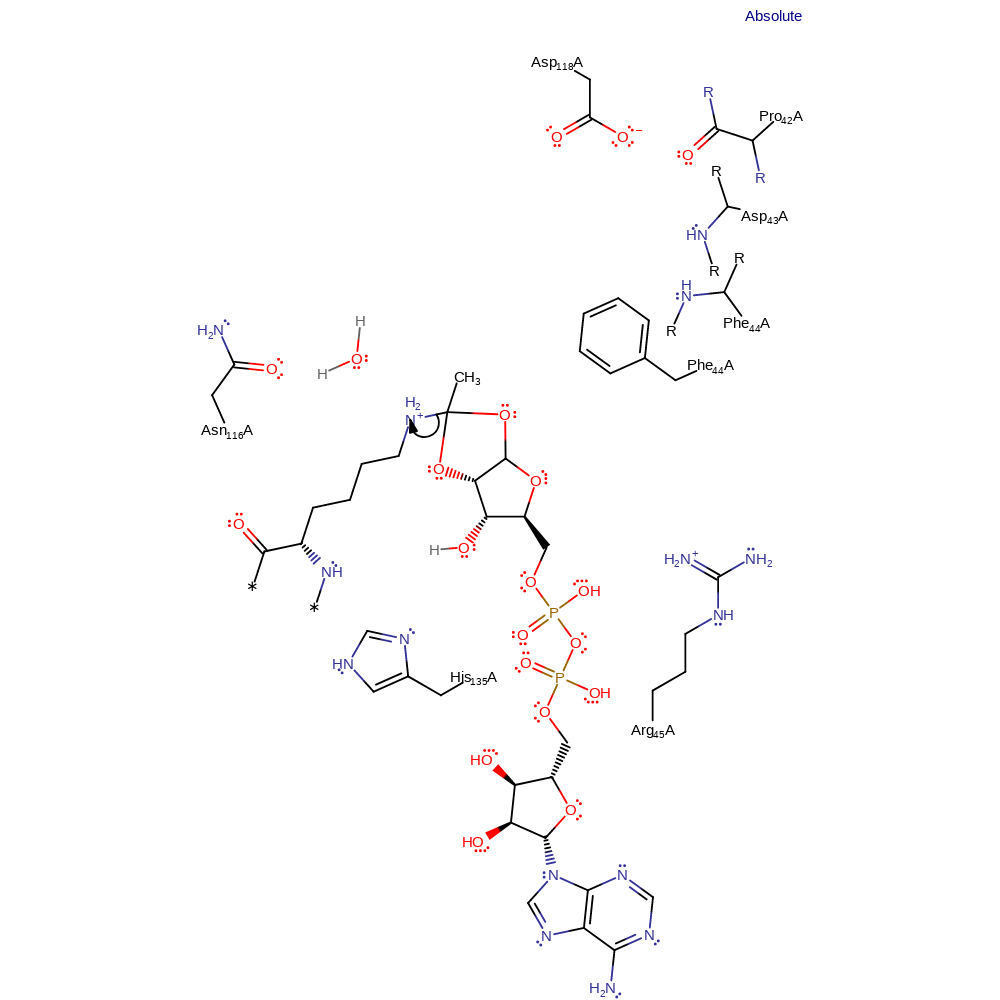
Step 5. Water, positioned and possibly activated by Asn116, attacks the carbocation.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Pro42(45)A (main-C) | hydrogen bond acceptor |
| Phe44(47)A (main-N) | van der waals interaction, hydrogen bond donor |
| Arg45(48)A | hydrogen bond donor |
| Asn116(119)A | increase nucleophilicity, hydrogen bond acceptor |
Chemical Components
ingold: bimolecular nucleophilic addition, intermediate formationCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Phe44(47)A (main-N) | van der waals interaction, hydrogen bond donor |
| Arg45(48)A | hydrogen bond donor |
| Asn116(119)A | hydrogen bond acceptor |
| His135(138)A | hydrogen bond acceptor |
Chemical Components
proton transfer, intermediate formation, overall product formed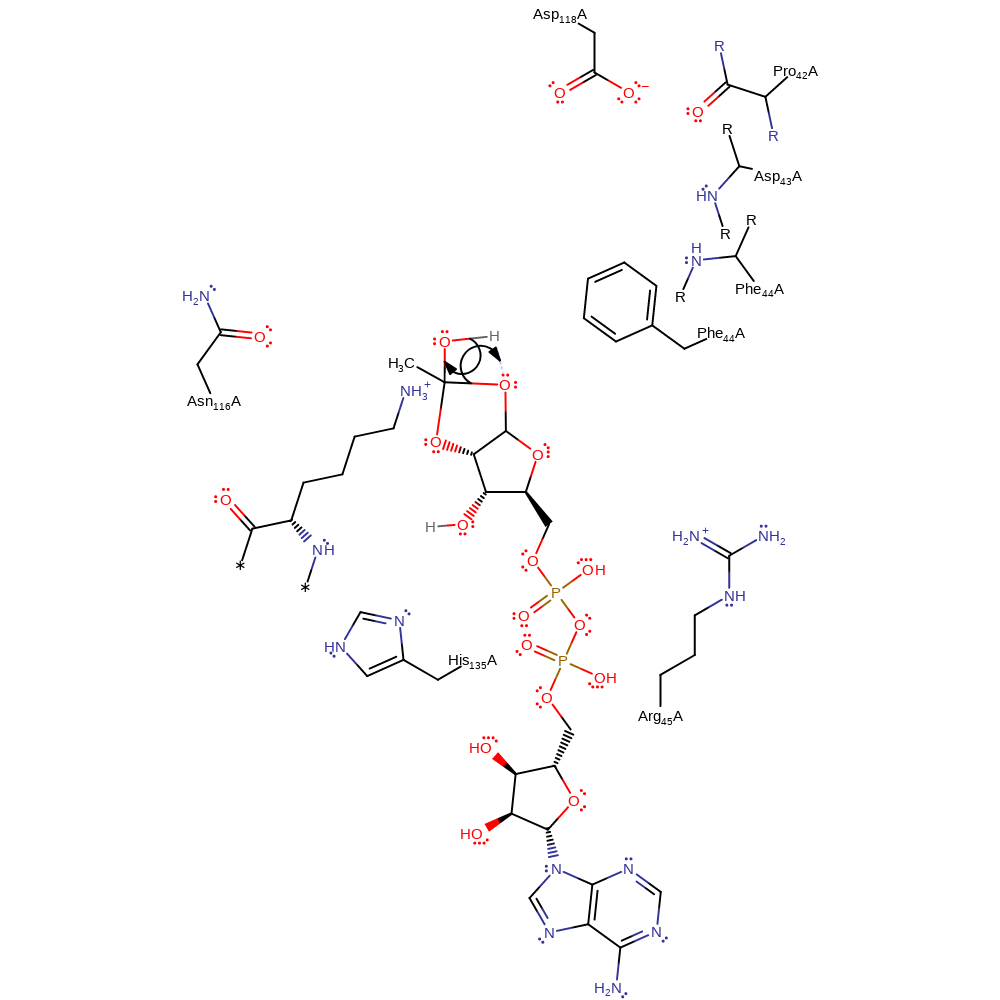
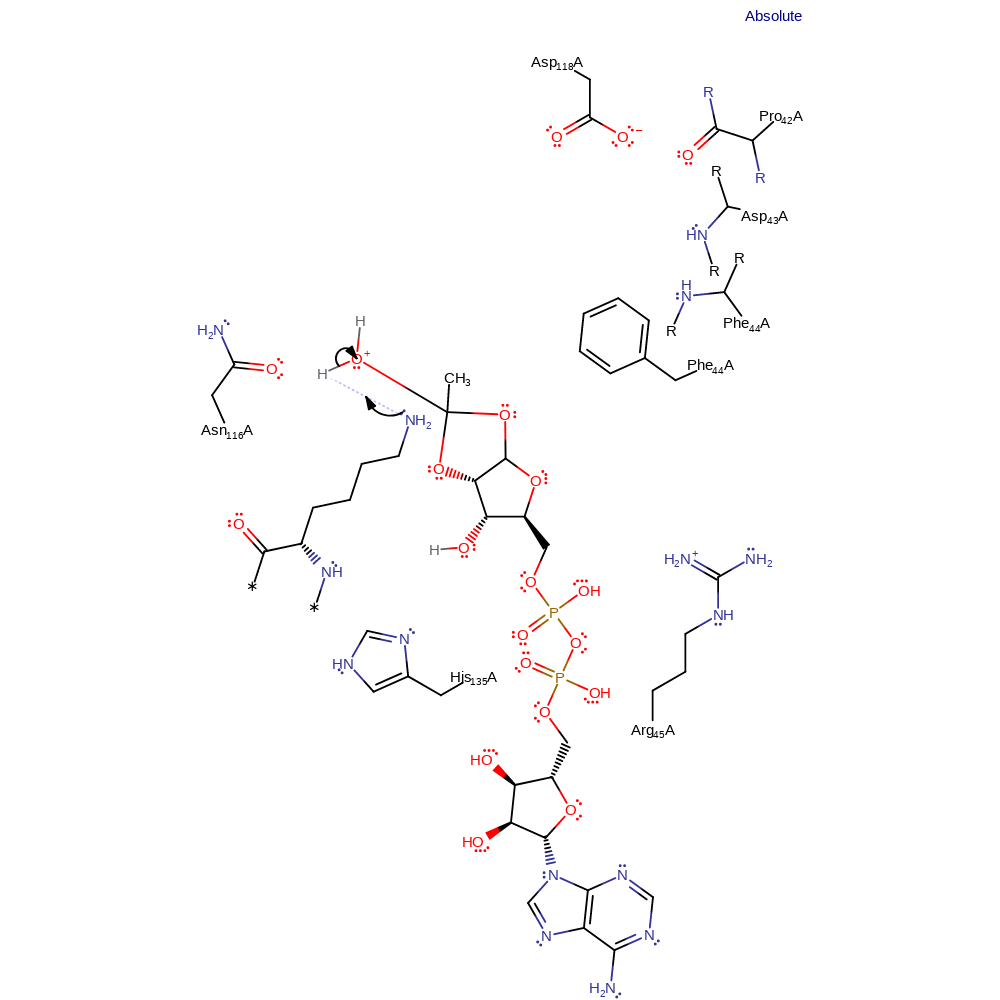
Step 7. The ring of the intermediate is opened when the exocyclic hydroxyl forms a carbonyl, with elimination and subsequent protonation of the endocyclic O1'.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Phe44(47)A (main-N) | van der waals interaction, hydrogen bond donor |
| Arg45(48)A | hydrogen bond donor |
| His135(138)A | hydrogen bond acceptor |






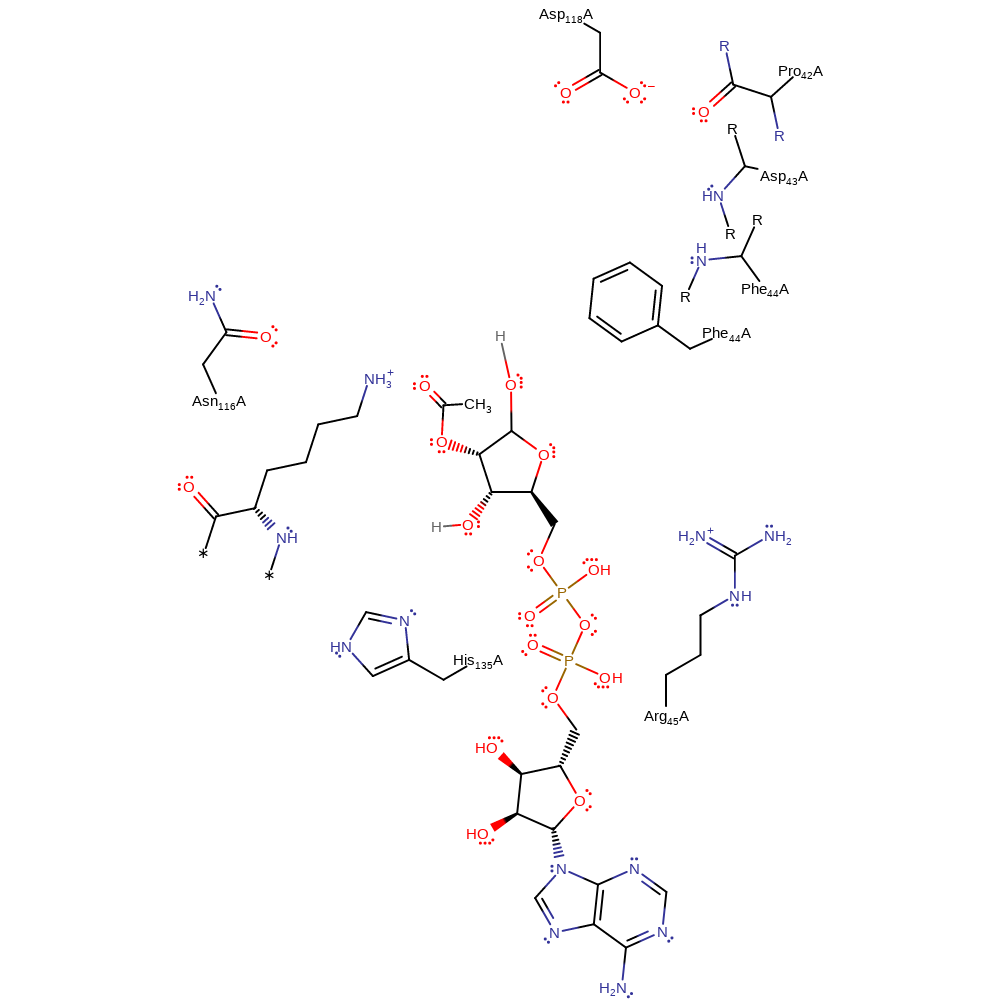 Download:
Download: