Arginine-tRNA ligase
Arginyl-tRNA synthetase (ArgRS), isolated from Saccharomyces cerevisiae, catalyses the esterification of arginine to the 3'-terminal adenosine of a tRNA molecule. The reaction proceeds via an aminoacyl-adenylate intermediate and is ATP dependent.
Although the tRNA molecule is not directly involved in the formation of the arginyl-adenylate intermediate, the tRNA must be bound to the enzyme before the reaction can occur. This may be due to conformational changes induced in the enzyme upon binding tRNA, as suggested for other aminoacyl-tRNA synthetases. ArgRS is a class 1a aminoacyl-tRNA synthease based on its structure, or class 1C when considering its requirement of tRNA.
Reference Protein and Structure
- Sequence
-
Q05506
 (6.1.1.19)
(6.1.1.19)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Saccharomyces cerevisiae S288c (Baker's yeast)

- PDB
-
1f7u
- CRYSTAL STRUCTURE OF THE ARGINYL-TRNA SYNTHETASE COMPLEXED WITH THE TRNA(ARG) AND L-ARG
(2.2 Å)



- Catalytic CATH Domains
-
3.40.50.620
 (see all for 1f7u)
(see all for 1f7u)
- Cofactors
- Magnesium(2+) (1) Metal MACiE
Enzyme Reaction (EC:6.1.1.19)
Enzyme Mechanism
Introduction
The carboxylate oxygen of arginine is the nucleophile for in-line attack on the alpha-phosphate of ATP. The transition state is trigonal bipyramidal and is stabilised by interactions with Lys156, His159, His162 and a magnesium ion (latter not found in crystal structures). The hydroxyl group of the 3'-terminal adenosine of tRNA is the nucleophile for attack on the ester of the resulting arginyl-adenylate, forming a tetrahedral intermediate. The intermediate collapses and eliminates AMP to form arginyl-tRNA.
Catalytic Residues Roles
| UniProt | PDB* (1f7u) | ||
| His159 | His159A(B) | His159 interacts with the alpha-phosphate of ATP and helps to stabilise the transition state for the formation of the arginyl-adenylate intermediate. It is part of the conserved HIGH motif. | hydrogen bond donor, electrostatic stabiliser |
| His162 | His162A(B) | His162 interacts with the ATP phosphates and so helps to stabilise the transition state for the formation of the arginyl-adenylate intermediate. | hydrogen bond donor, electrostatic stabiliser |
| Lys156 | Lys156A(B) | Lys156 forms a salt bridge with one of the non-bridging oxygens of the alpha-phosphate of ATP. This stabilises the transition states of aminoacylation, in particular the formation of the arginyl-adenylate intermediate. | attractive charge-charge interaction, hydrogen bond donor, electrostatic stabiliser |
Chemical Components
bimolecular nucleophilic substitution, intermediate formation, overall reactant used, overall product formed, bimolecular nucleophilic addition, proton transfer, intramolecular elimination, native state of enzyme regenerated, intermediate collapse, intermediate terminatedReferences
- Sekine S et al. (2001), J Biol Chem, 276, 3723-3726. Crucial Role of the HIGH-loop Lysine for the Catalytic Activity of Arginyl-tRNA Synthetase. DOI:10.1074/jbc.c000756200. PMID:11106639.
- Konno M et al. (2009), FEBS J, 276, 4763-4779. Modeling of tRNA-assisted mechanism of Arg activation based on a structure of Arg-tRNA synthetase, tRNA, and an ATP analog (ANP). DOI:10.1111/j.1742-4658.2009.07178.x. PMID:19656186.
- Airas RK (2006), Biochim Biophys Acta, 1764, 307-319. Analysis of the kinetic mechanism of arginyl-tRNA synthetase. DOI:10.1016/j.bbapap.2005.11.020. PMID:16427818.
- Sekine S et al. (2003), EMBO J, 22, 676-688. ATP binding by glutamyl-tRNA synthetase is switched to the productive mode by tRNA binding. DOI:10.1093/emboj/cdg053. PMID:12554668.
- Delagoutte B et al. (2000), EMBO J, 19, 5599-5610. tRNA aminoacylation by arginyl-tRNA synthetase: induced conformations during substrates binding. DOI:10.1093/emboj/19.21.5599. PMID:11060012.
- Cavarelli J et al. (1998), EMBO J, 17, 5438-5448. L-Arginine recognition by yeast arginyl-tRNA synthetase. DOI:10.1093/emboj/17.18.5438. PMID:9736621.
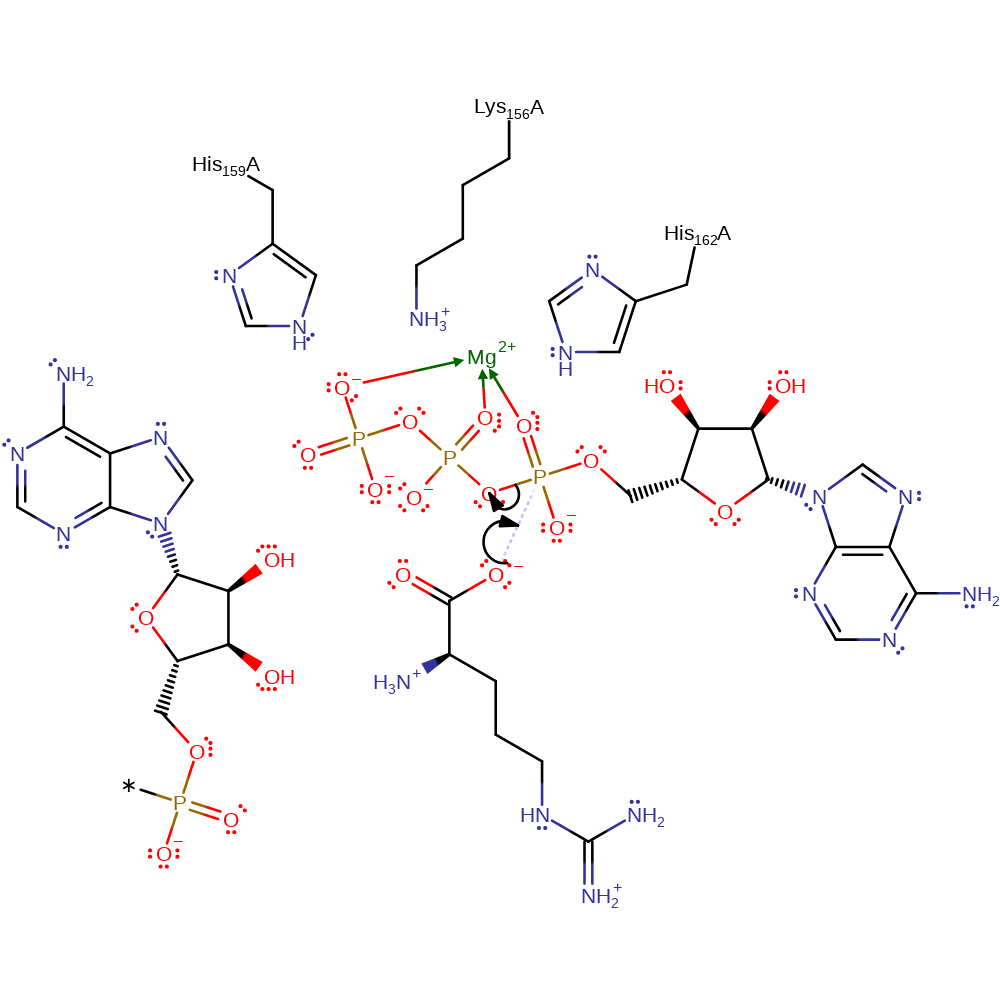
Step 1. The carboxylate oxygen of arginine is the nucleophile for in-line displacement at the alpha-phosphate of ATP. This produces pyrophosphate and an arginyl-adenylate intermediate.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His162A(B) | electrostatic stabiliser, hydrogen bond donor |
| His159A(B) | hydrogen bond donor, electrostatic stabiliser |
| Lys156A(B) | hydrogen bond donor, attractive charge-charge interaction, electrostatic stabiliser |
Chemical Components
ingold: bimolecular nucleophilic substitution, intermediate formation, overall reactant used, overall product formed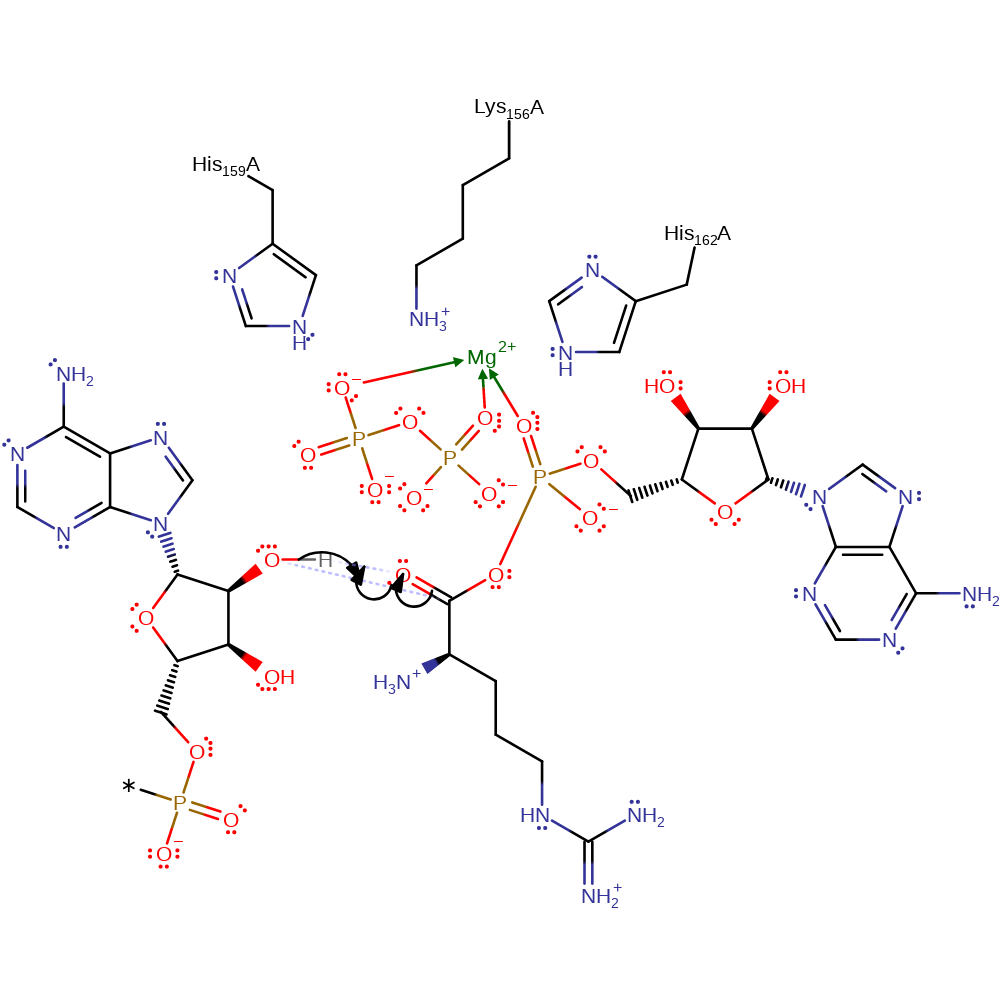
Step 2. The 2'-hydroxyl of the 3'-terminal adenosine of the tRNA molecule is the nucleophile for attack on the carbonyl carbon of the arginyl-adenylate intermediate. This forms a tetrahedral intermediate.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|
Chemical Components
ingold: bimolecular nucleophilic addition, proton transfer, intermediate formation, overall reactant used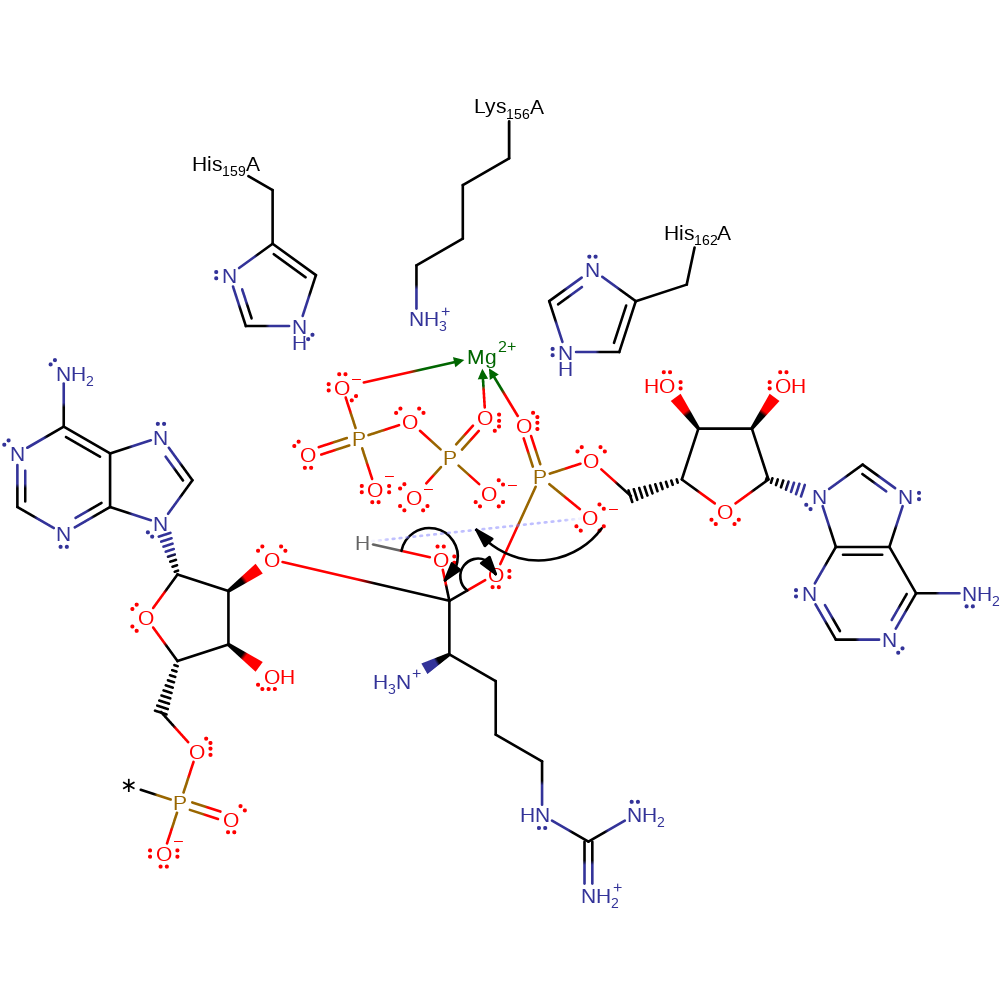
Step 3. The tetrahedral intermediate collapses and eliminates AMP to form L-arginyl-tRNA.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|






 Download:
Download: