NAD+ synthase
NAD+ is involved electron transport and redox reactions and in DNA ligation and protein ADP-ribosylation. In yeast and most other organisms, NAD is generated through the de novo pathway and the salvage pathway. In the de novo pathway, quinolinic acid is converted to nicotinic acid mononucleotide (NaMN). In the salvage pathway, NaMN is generated by recycling of nicotinamide. Both pathways converge on NaMN, which is then converted into deamido-NAD+. Subsequently, deamido-NAD+ is converted to NAD+ by NAD+ synthetase. This entry represents NH(3)-dependent NAD(+) synthetases from prokaryotes.
Reference Protein and Structure
- Sequence
-
P08164
 (6.3.1.5)
(6.3.1.5)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Bacillus subtilis subsp. subtilis str. 168 (Bacteria)

- PDB
-
1kqp
- NH3-DEPENDENT NAD+ SYNTHETASE FROM BACILLUS SUBTILIS AT 1 A RESOLUTION
(1.03 Å)



- Catalytic CATH Domains
-
3.40.50.620
 (see all for 1kqp)
(see all for 1kqp)
- Cofactors
- Magnesium(2+) (2), Water (2) Metal MACiE
Enzyme Reaction (EC:6.3.1.5)
Enzyme Mechanism
Introduction
There is no direct involvement in this mechanism by any amino acid residues in the active site. The carboxylate group of the deamino-NAD acts as a nucleophile and attacks the gamma-phosphate of the ATP in a substitution reaction, liberating pyrophosphate. The carbonyl group of the phosphorylated substrate accepts a proton from the ammonium ion, thus increasing the electrophilicity of the carbon atom. Water deprotonates the ammonia molecule, which then acts as a nucleophile and attacks the carboxyl carbon of the phosphorylated intermediate in a substitution reaction, liberating AMP. A second water molecule deprotonates the alcohol group formed in the previous step, regenerating the carbonyl group.
Catalytic Residues Roles
| UniProt | PDB* (1kqp) | ||
| Asp51, Glu163 | Asp50A, Glu162A | Form part of the magensium binding site. | metal ligand |
Chemical Components
bimolecular nucleophilic substitution, overall reactant used, intermediate formation, overall product formed, proton transfer, intermediate terminatedReferences
- Symersky J et al. (2002), Acta Crystallogr D Biol Crystallogr, 58, 1138-1146. NH3-dependent NAD+ synthetase from Bacillus subtilis at 1 A resolution. DOI:10.2210/pdb1kqp/pdb. PMID:12077433.
- De Ingeniis J et al. (2012), PLoS One, 7, e39115-. Glutamine versus ammonia utilization in the NAD synthetase family. DOI:10.1371/journal.pone.0039115. PMID:22720044.
- Devedjiev Y et al. (2001), Acta Crystallogr D Biol Crystallogr, 57, 806-812. Stabilization of active-site loops in NH3-dependent NAD+synthetase fromBacillus subtilis. DOI:10.1107/s0907444901003523. PMID:11375500.
- Rizzi M et al. (1998), Structure, 6, 1129-1140. A novel deamido-NAD+-binding site revealed by the trapped NAD-adenylate intermediate in the NAD+ synthetase structure. DOI:10.1016/s0969-2126(98)00114-2. PMID:9753692.
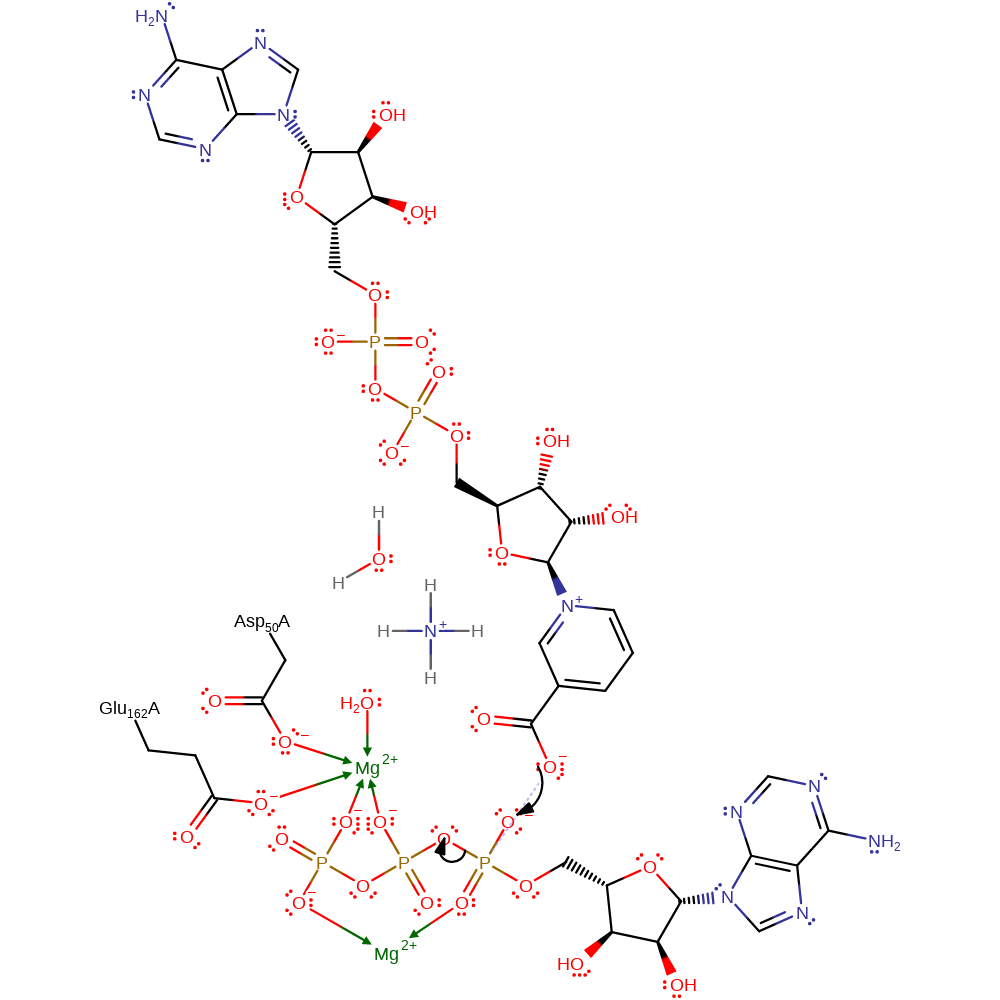
Step 1. Carboxylate group of the deamino-NAD acts as a nucleophile and attacks the gamma-phosphate of the ATP in a substitution reaction, liberating pyrophosphate. The two Mg(II) ions stabilise the intermediates formed
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu162A | metal ligand |
| Asp50A | metal ligand |
Chemical Components
ingold: bimolecular nucleophilic substitution, overall reactant used, intermediate formation, overall product formed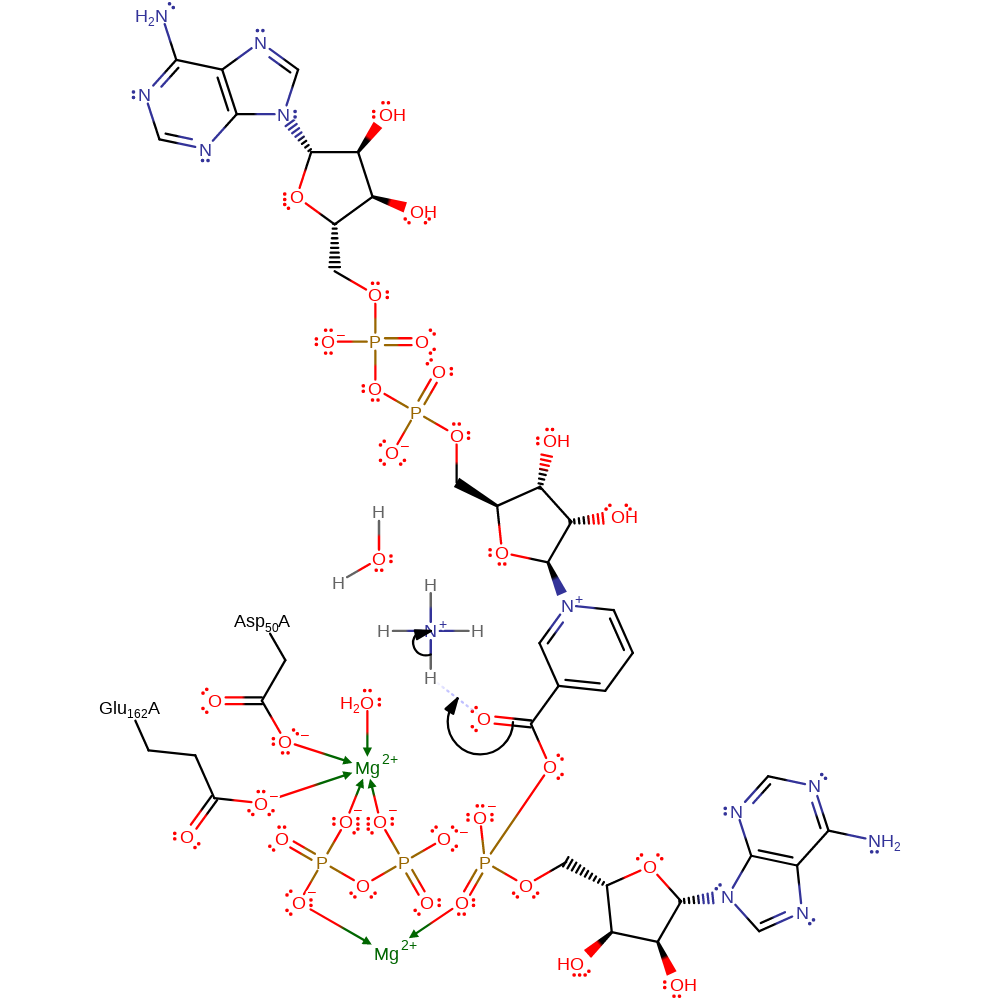
Step 2. The carbonyl group of the phosphorylated substrate accepts a proton from the ammonium ion, thus increasing the electrophilicity of the carbon atom.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu162A | metal ligand |
| Asp50A | metal ligand |
Chemical Components
proton transfer, overall reactant used, intermediate formation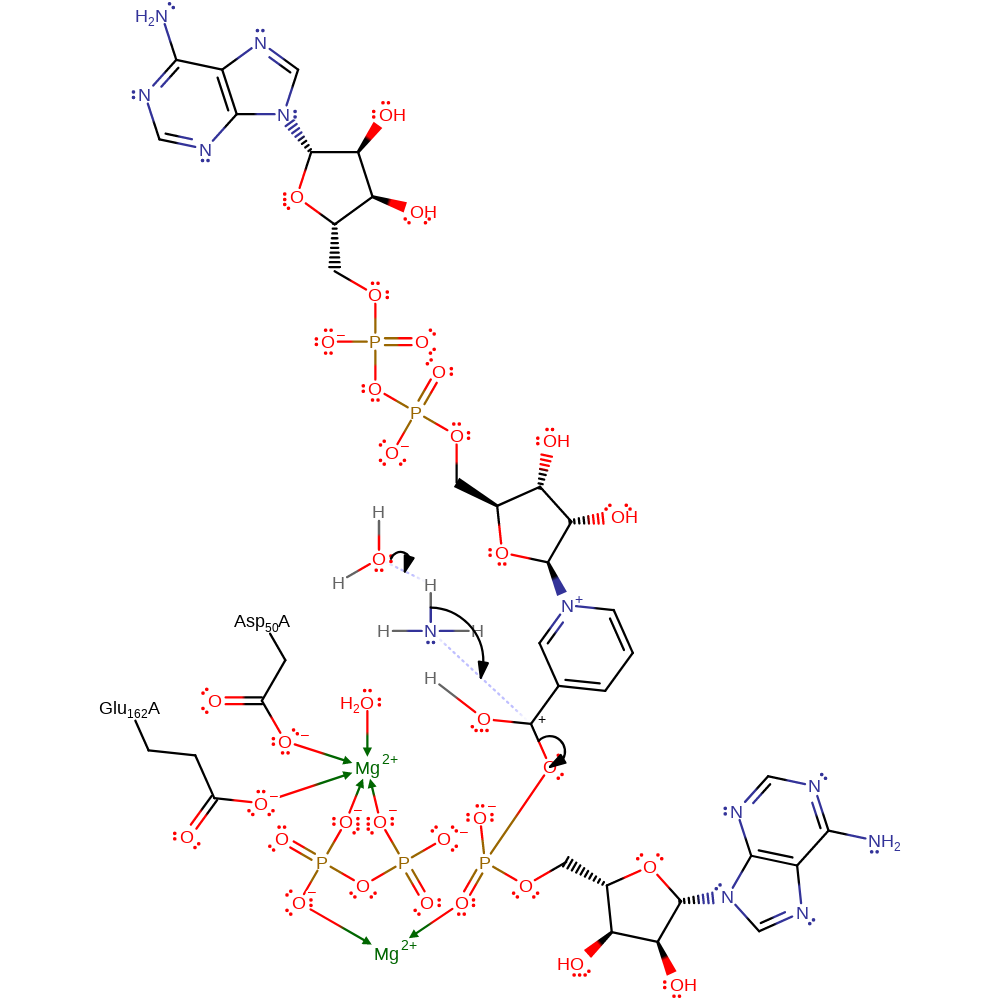
Step 3. Water deprotonates the ammonia molecule, which then acts as a nucleophile and attacks the carboxyl carbon of the phosphorylated intermediate in a substitution reaction, liberating AMP.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu162A | metal ligand |
| Asp50A | metal ligand |
Chemical Components
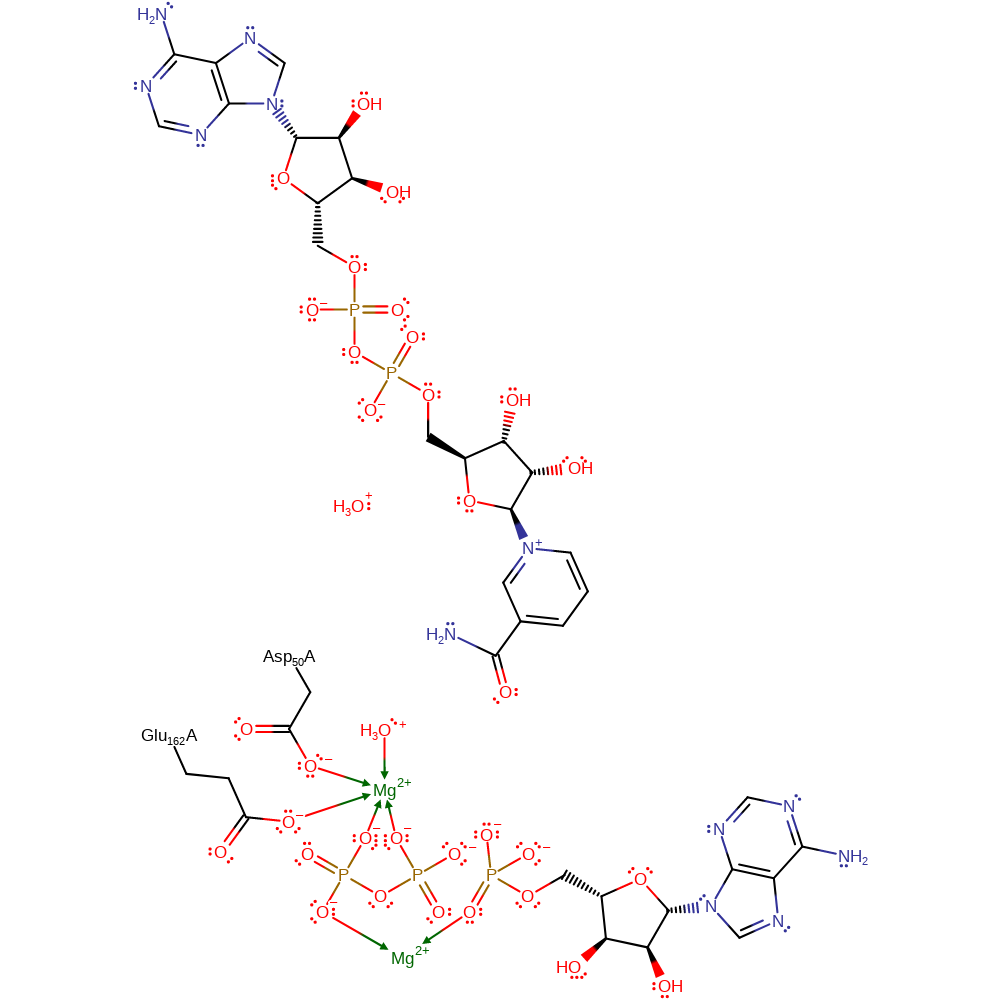
proton transfer, ingold: bimolecular nucleophilic substitution, intermediate terminated, overall product formed
Step 4. A second water molecule deprotonates the alcohol group formed in the previous step, regenerating the carbonyl group.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu162A | metal ligand |
| Asp50A | metal ligand |







 Download:
Download: