Trypsin
The mechanism of the serine proteases is commonly believed to be among the best understood of all enzymes. As their name suggests, they are involved in hydrolysis of proteins using a serine nucleophile. As well as the active site, there is also a 'specificity pocket' which determines which amino acids the enzyme will cleave at. For trypsin, this pocket contains a negatively charged residue, which results in it having a preference for cleaving at positively charged residues i.e. lysine or arginine.
Reference Protein and Structure
- Sequence
-
P35049
 (3.4.21.4)
(3.4.21.4)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Fusarium oxysporum (Fungus)

- PDB
-
1pq5
- Trypsin at pH 5, 0.85 A
(0.85 Å)



- Catalytic CATH Domains
-
2.40.10.10
 (see all for 1pq5)
(see all for 1pq5)
Enzyme Reaction (EC:3.4.21.4)
Enzyme Mechanism
Introduction
The key feature of the mechanism is the presence of the catalytic triad of serine, histidine and aspartate. Serine, having been deprotonated by histidine, attacks the carbonyl of the substrate. The negatively charged tetrahedral intermediate is stabilised by the oxyanion hole, while the positive charge on histidine is stabilised by the aspartate residue. When the tetrahedral intermediate collapses, the amide bond of the substrate is broken. The acylenzyme intermediate is hydrolysed by a water molecule, activated by histidine, to release the product and restore the enzyme to its active state.
Catalytic Residues Roles
| UniProt | PDB* (1pq5) | ||
| His65 | His56(41)A | Forms part of the catalytic triad. Deprotonates Ser195 to activate it as a nucleophile, and is stabilised by hydrogen bonding interactions with Asp99. In the deacylation step, deprotonates a water molecule to activate it as a nucleophile. | hydrogen bond acceptor, hydrogen bond donor, proton acceptor, proton donor |
| Asp108 | Asp99(84)A | Forms part of the catalytic triad. Stabilises the positive charge on His56. | activator, hydrogen bond acceptor, electrostatic stabiliser |
| Ser204 | Ser195(180)A | When activated by His56, Ser195 is the nucleophile which attacks the substrate carbonyl. | covalently attached, hydrogen bond acceptor, hydrogen bond donor, nucleofuge, proton acceptor, proton donor, nucleophile, electrostatic stabiliser |
| Gly202 (main-N), Ser204 (main-N), Gln201 (main-N), Asp203 (main-N) | Gly193(178)A (main-N), Ser195(180)A (main-N), Gln192(177)A (main-N), Asp194(179)A (main-N) | Forms part of the oxyanion hole which polarises the substrate carbonyl to facilitate attack and then stabilises the negatively charged oxygen in the transition state. | hydrogen bond donor, electrostatic stabiliser, transition state stabiliser |
Chemical Components
proton transfer, bimolecular nucleophilic addition, overall reactant used, enzyme-substrate complex formation, intermediate formation, unimolecular elimination by the conjugate base, enzyme-substrate complex cleavage, intermediate collapse, overall product formed, intermediate terminated, native state of enzyme regeneratedReferences
- Schmidt A et al. (2003), J Biol Chem, 278, 43357-43362. Trypsin Revisited: CRYSTALLOGRAPHY AT (SUB) ATOMIC RESOLUTION AND QUANTUM CHEMISTRY REVEALING DETAILS OF CATALYSIS. DOI:10.1074/jbc.m306944200. PMID:12937176.
- Blankenship E et al. (2014), Acta Crystallogr D Biol Crystallogr, 70, 833-840. Conformational flexibility in the catalytic triad revealed by the high-resolution crystal structure of Streptomyces erythraeus trypsin in an unliganded state. DOI:10.1107/S1399004713033658. PMID:24598752.
- Wahlgren WY et al. (2011), J Biol Chem, 286, 3587-3596. The catalytic aspartate is protonated in the Michaelis complex formed between trypsin and an in vitro evolved substrate-like inhibitor: a refined mechanism of serine protease action. DOI:10.1074/jbc.M110.161604. PMID:21097875.
- Topf M et al. (2002), Proteins, 47, 357-369. Molecular dynamics simulations of the acyl-enzyme and the tetrahedral intermediate in the deacylation step of serine proteases. DOI:10.1002/prot.10097. PMID:11948789.

Step 1. His56 in a Ser-His-Asp triad deprotonates Ser195. Activated Ser195 then attacks the carbonyl carbon of the peptide bond in a nucleophilic addition.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp99(84)A | hydrogen bond acceptor, activator |
| His56(41)A | hydrogen bond acceptor, hydrogen bond donor |
| Ser195(180)A | hydrogen bond donor |
| Asp194(179)A (main-N) | hydrogen bond donor |
| Gly193(178)A (main-N) | hydrogen bond donor |
| Gln192(177)A (main-N) | hydrogen bond donor, transition state stabiliser |
| Gly193(178)A (main-N) | transition state stabiliser |
| Asp194(179)A (main-N) | transition state stabiliser |
| Ser195(180)A (main-N) | transition state stabiliser |
| Ser195(180)A | proton donor, nucleophile |
| His56(41)A | proton acceptor |
Chemical Components
proton transfer, ingold: bimolecular nucleophilic addition, overall reactant used, enzyme-substrate complex formation, intermediate formation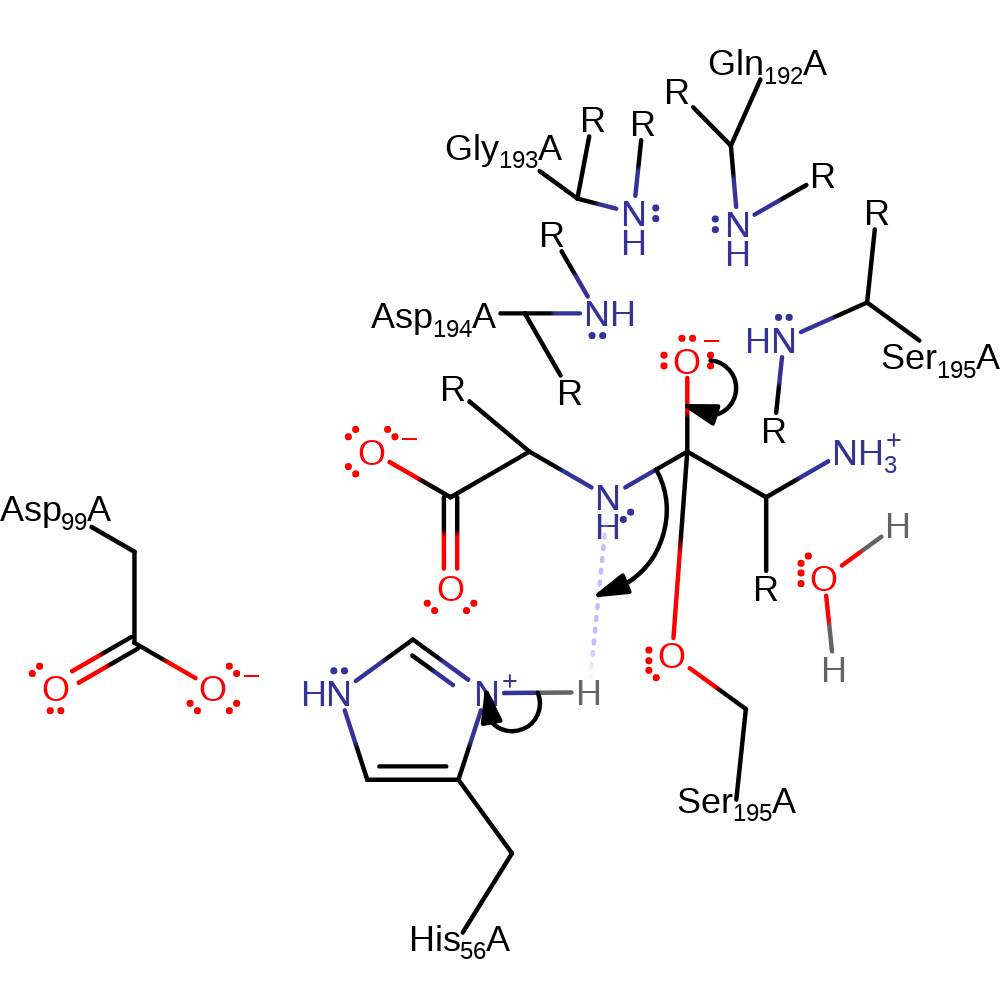
Step 2. The oxyanion initiates an elimination reaction that cleaves the peptide bond, releasing the new N-terminus of the protein, which protonates from His56.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp99(84)A | hydrogen bond acceptor, electrostatic stabiliser |
| His56(41)A | hydrogen bond donor |
| Ser195(180)A | covalently attached, hydrogen bond acceptor, hydrogen bond donor, electrostatic stabiliser |
| Asp194(179)A (main-N) | hydrogen bond donor, electrostatic stabiliser |
| Gly193(178)A (main-N) | hydrogen bond donor, electrostatic stabiliser |
| Gln192(177)A (main-N) | hydrogen bond donor, electrostatic stabiliser |
| Ser195(180)A (main-N) | electrostatic stabiliser |
| His56(41)A | proton donor |
Chemical Components
proton transfer, ingold: unimolecular elimination by the conjugate base, enzyme-substrate complex cleavage, intermediate collapse, intermediate formation, overall product formed
Step 3. His56 deprotonates water, which attacks the carbonyl carbon bound to Ser195 in a nucleophilic addition.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp99(84)A | hydrogen bond acceptor |
| His56(41)A | hydrogen bond donor, hydrogen bond acceptor |
| Ser195(180)A | covalently attached, hydrogen bond donor |
| Asp194(179)A (main-N) | hydrogen bond donor |
| Gly193(178)A (main-N) | hydrogen bond donor |
| Gln192(177)A (main-N) | hydrogen bond donor, transition state stabiliser |
| Gly193(178)A (main-N) | transition state stabiliser |
| Asp194(179)A (main-N) | transition state stabiliser |
| Ser195(180)A (main-N) | transition state stabiliser |
| His56(41)A | proton acceptor |
Chemical Components
proton transfer, ingold: bimolecular nucleophilic addition, overall reactant used, enzyme-substrate complex formation, intermediate formation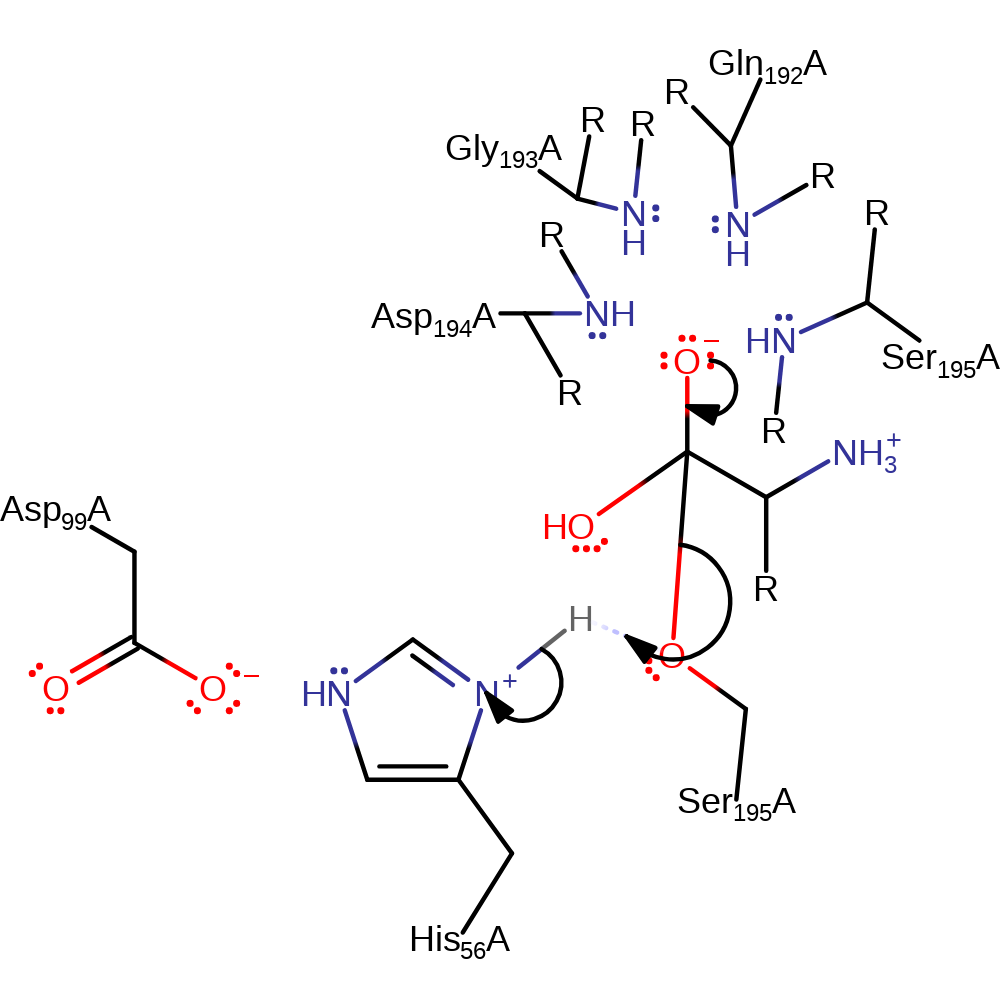
Step 4. The oxyanion initiates an elimination that cleaves the acyl bond to Ser195, releasing the C-terminus of the protein. Ser195 then deprotonates His56, regenerating the active site.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp99(84)A | hydrogen bond acceptor, electrostatic stabiliser |
| His56(41)A | hydrogen bond donor |
| Ser195(180)A | hydrogen bond acceptor, hydrogen bond donor, electrostatic stabiliser |
| Asp194(179)A (main-N) | hydrogen bond donor, electrostatic stabiliser |
| Gly193(178)A (main-N) | hydrogen bond donor, electrostatic stabiliser |
| Gln192(177)A (main-N) | hydrogen bond donor, electrostatic stabiliser |
| Ser195(180)A (main-N) | electrostatic stabiliser |
| His56(41)A | proton donor |
| Ser195(180)A | nucleofuge, proton acceptor |



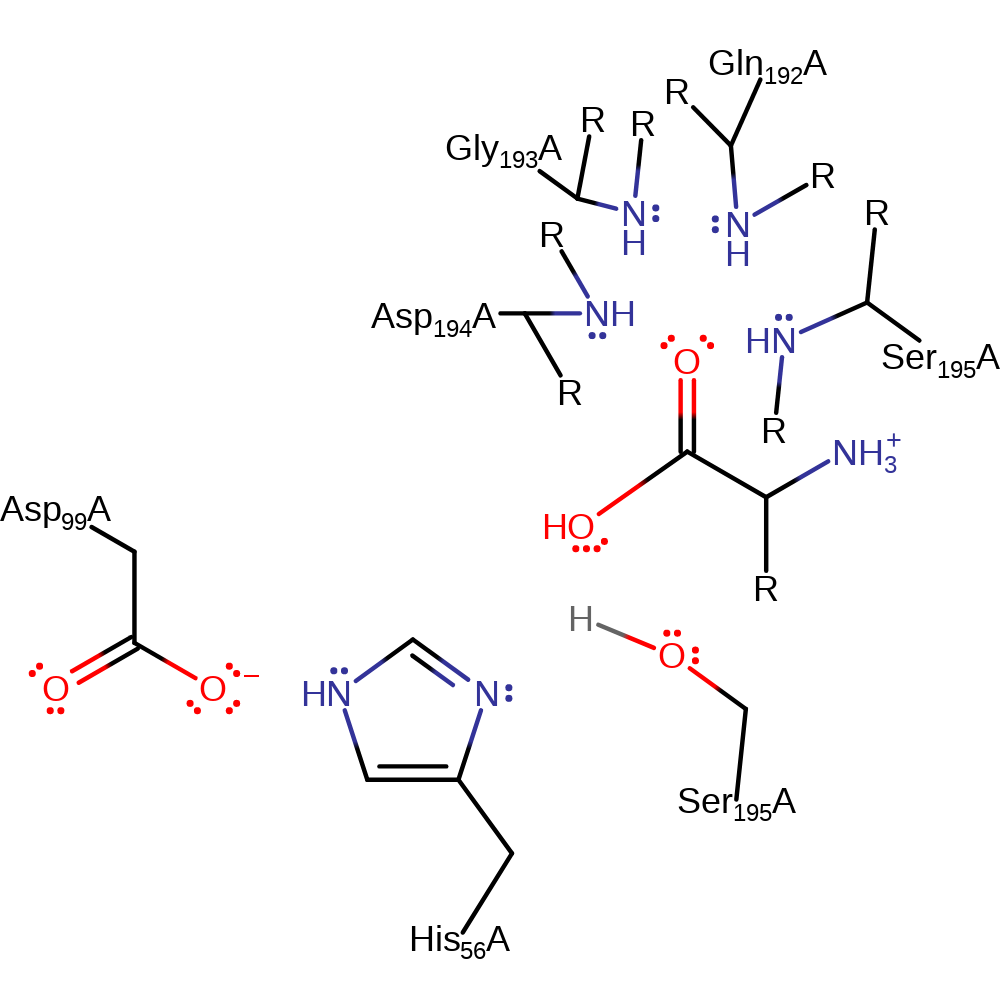 Download:
Download: