Deacetoxycephalosporin-C synthase
Deacetoxycephalosporin C synthase (DAOCS) from Streptomyces clavuligerus is able to catalyse the synthesis of sporins from penicillin using 2-oxoglutarate (2OG) as a co-substrate. This reaction is potentially very useful to industry because it enables processing of penicillin to other beta-lactam ring containing compounds which may not trigger antibiotic resistance systems in prokaryotes that evolved with penicillin itself as the selective agent. As with the other members of the family of 2OG utilising enzymes, DAOCS shows a beta barrel catalytic core and has its iron centre ligated to two histidines and an aspartate residue at the active site. It also displays roughly the same catalytic cycle, albeit for a specific reaction, as seen for the other members of the family, such as anthocyanidin synthase.
Reference Protein and Structure
- Sequence
-
P18548
 (1.14.20.1)
(1.14.20.1)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Streptomyces clavuligerus (Bacteria)

- PDB
-
1unb
- Deacetoxycephalosporin C synthase complexed with 2-oxoglutarate and ampicillin
(1.5 Å)



- Catalytic CATH Domains
-
2.60.120.330
 (see all for 1unb)
(see all for 1unb)
- Cofactors
- Iron(2+) (1) Metal MACiE
Enzyme Reaction (EC:1.14.20.1)
Enzyme Mechanism
Introduction
There are two half reactions that occur in the enzyme which are independent in as much as they can be uncoupled under certain conditions. In the oxidative half reaction the Fe (II) centre binds to dioxygen, acting as a nucleophile thus becoming oxidised to Fe (III). This creates a peroxide radical which reacts with the 2OG bound to the iron causing decarboxylation and the further oxidation of iron to the highly reactive Fe (IV) species with a double bond to oxygen. In the reductive half reaction the double bonded oxygen accepts a hydrogen atom from the beta methyl group of penicillin to form an OH group liganded to the iron (now Fe (III)) and an alkyl radical. The alkyl radical reacts with the sulphur of the ring to cause the expansion of the ring and the migration of the radical electron to the 2C of the ring. From here, hydrogen transfer to the OH group via the Sulphur atom occurs leading to the formation of a double bond between the 2C and the 3C of the ring, and the formation of Fe (II) bonded to a water molecule. This completes the reaction cycle. Arg 74 ensures that the hydrogen transfer can occur by sterically constraining penicillin so that it is in the same plane as the Fe (IV) double bonded oxygen.
Catalytic Residues Roles
| UniProt | PDB* (1unb) | ||
| Arg74 | Arg74A | Sterically constrains the penicillin substrate thus positioning it in the optimal position for hydrogen transfer. | steric hindrance |
Chemical Components
bimolecular electrophilic addition, coordination, overall reactant used, coordination to a metal ion, intermediate formation, unimolecular elimination by the conjugate base, intermediate collapse, overall product formed, decarboxylation, hydrogen transfer, radical formation, redox reaction, colligation, radical propagation, cyclisation, homolysis, decyclisation, electron transfer, proton transfer, radical termination, heterolysis, decoordination from a metal ion, intermediate terminated, native state of enzyme regeneratedReferences
- Clifton IJ et al. (2006), J Inorg Biochem, 100, 644-669. Structural studies on 2-oxoglutarate oxygenases and related double-stranded β-helix fold proteins. DOI:10.1016/j.jinorgbio.2006.01.024. PMID:16513174.
- Valegård K et al. (2004), Nat Struct Mol Biol, 11, 95-101. The structural basis of cephalosporin formation in a mononuclear ferrous enzyme. DOI:10.1038/nsmb712. PMID:14718929.
- Oster LM et al. (2004), J Mol Biol, 343, 157-171. Conformational Flexibility of the C Terminus with Implications for Substrate Binding and Catalysis Revealed in a New Crystal Form of Deacetoxycephalosporin C Synthase. DOI:10.1016/j.jmb.2004.07.049. PMID:15381427.
- Lee HJ et al. (2001), J Mol Biol, 308, 937-948. Kinetic and crystallographic studies on deacetoxycephalosporin C synthase (DAOCS). DOI:10.1006/jmbi.2001.4649. PMID:11352583.
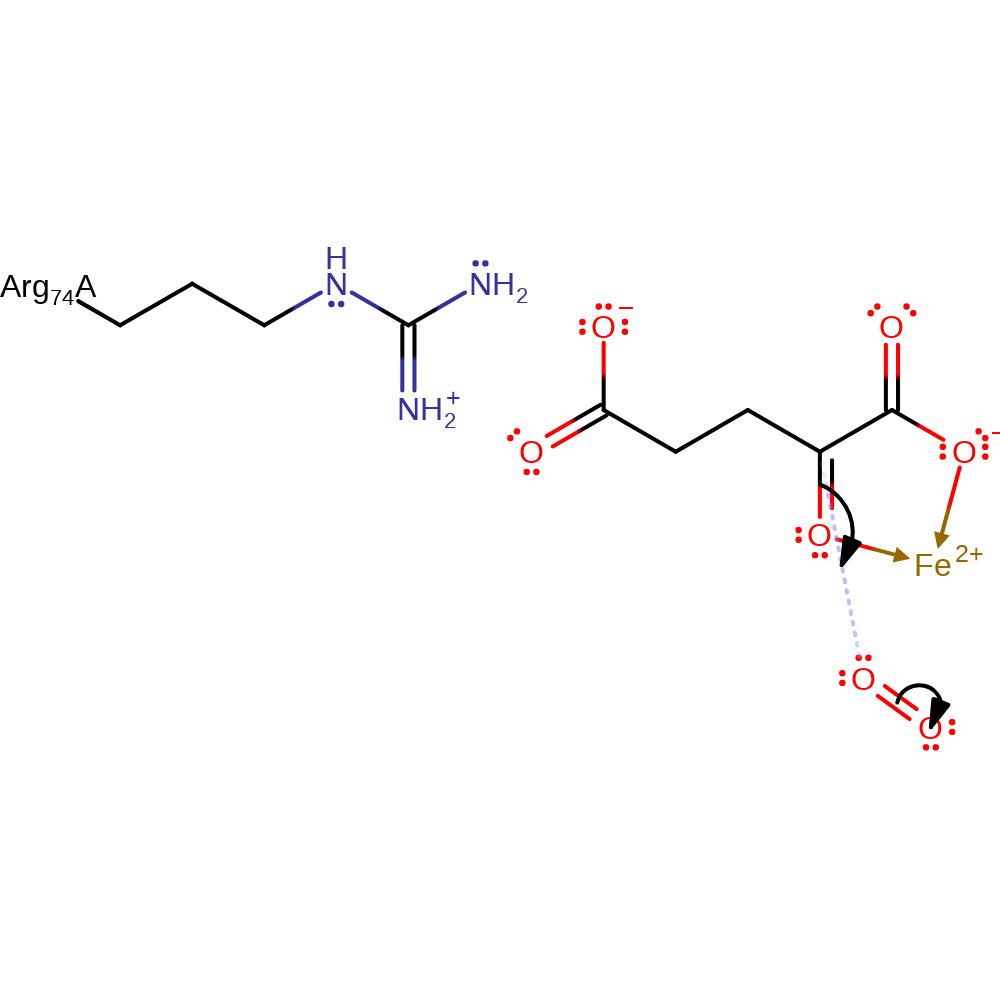
Step 1. 2-oxoglutarate initiates an electrophilic addition to dioxygen, which coordinates to the Fe(II) cofactor.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Arg74A | steric hindrance |
Chemical Components
ingold: bimolecular electrophilic addition, coordination, overall reactant used, coordination to a metal ion, intermediate formation
Step 2. Carbon dioxide is eliminated from the intermediate to form a planar peroxo intermediate. Carbon dioxide is still coordinated to the Fe(II) cofactor.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Arg74A | steric hindrance |
Chemical Components
ingold: unimolecular elimination by the conjugate base, intermediate collapse, intermediate formation, overall product formed, decarboxylation
Step 3. Fe(II) initiates an elimination reaction by donating two electrons to the bound peroxo moiety, cleaving the peroxo bond and producing the succinate product and iron as a ferryl species.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Arg74A | steric hindrance |
Chemical Components
ingold: unimolecular elimination by the conjugate base, intermediate formation, overall product formed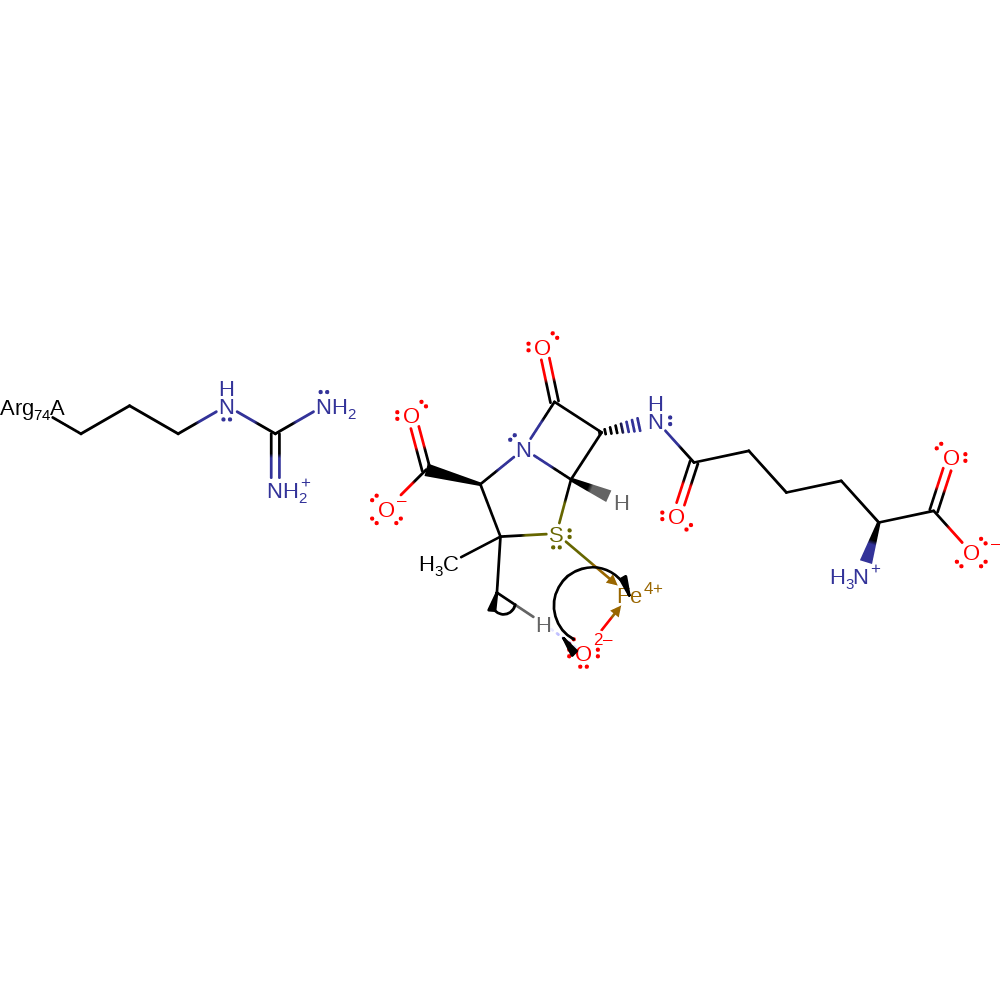
Step 4. Penicillin binds, displacing the succinate and carbon dioxide as iron ligands. The oxo group deprotonates the CH3 of the penicillin, generating a alkyl radical and resulting in the donation of a single electron to Fe(IV).
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Arg74A | steric hindrance |
Chemical Components
hydrogen transfer, radical formation, redox reaction, overall reactant used, intermediate formation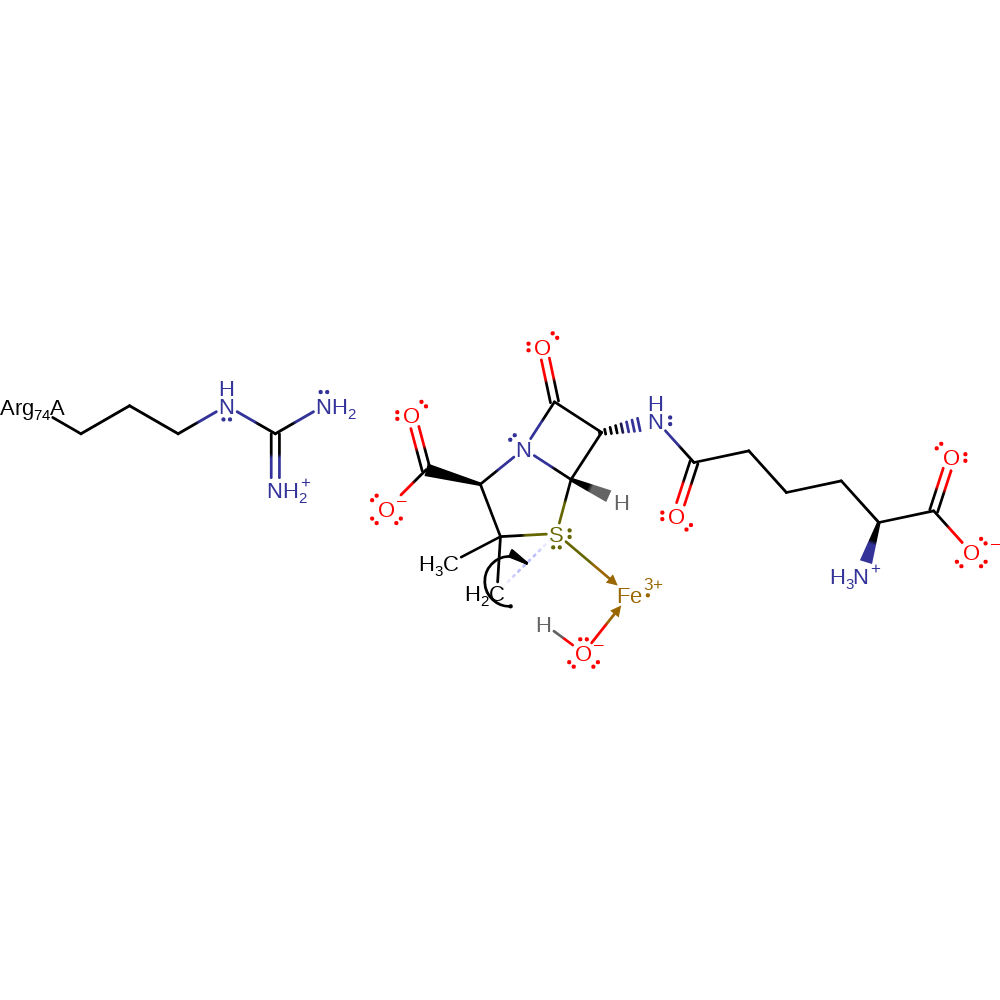
Step 5. A colligation occurs between the alkyl radical and the sulfur of the five membered ring in the penicillin.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Arg74A | steric hindrance |
Chemical Components
colligation, radical propagation, intermediate formation, cyclisation
Step 6. The C-S bond of the original five-mebered ring homolyses, producing the six-membered ring.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Arg74A | steric hindrance |
Chemical Components
homolysis, radical propagation, intermediate formation, decyclisation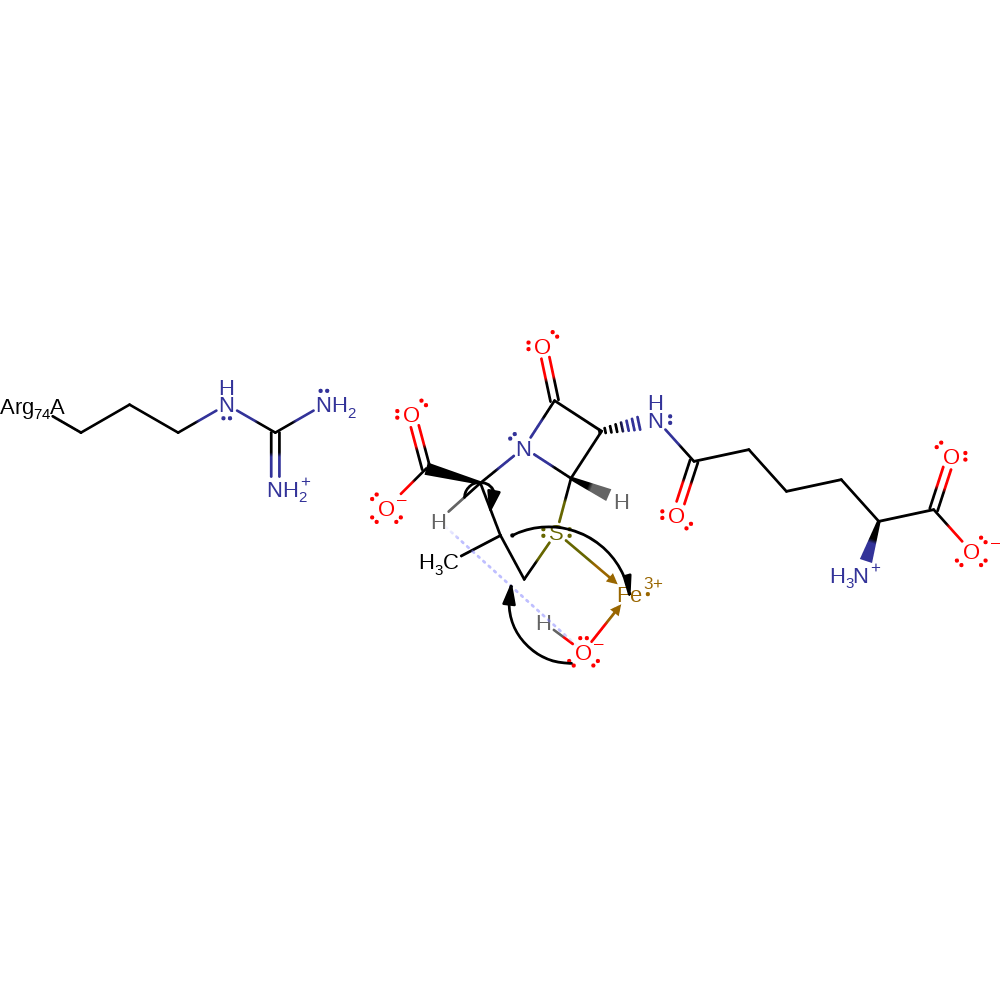
Step 7. The carbon radical in the ring donates a single electron to the Fe(III) cofactor, which causes the bound hydroxide to deprotonate the C3 of the thiazolidine ring generating the final deacetoxycephalosporin C product.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Arg74A | steric hindrance |







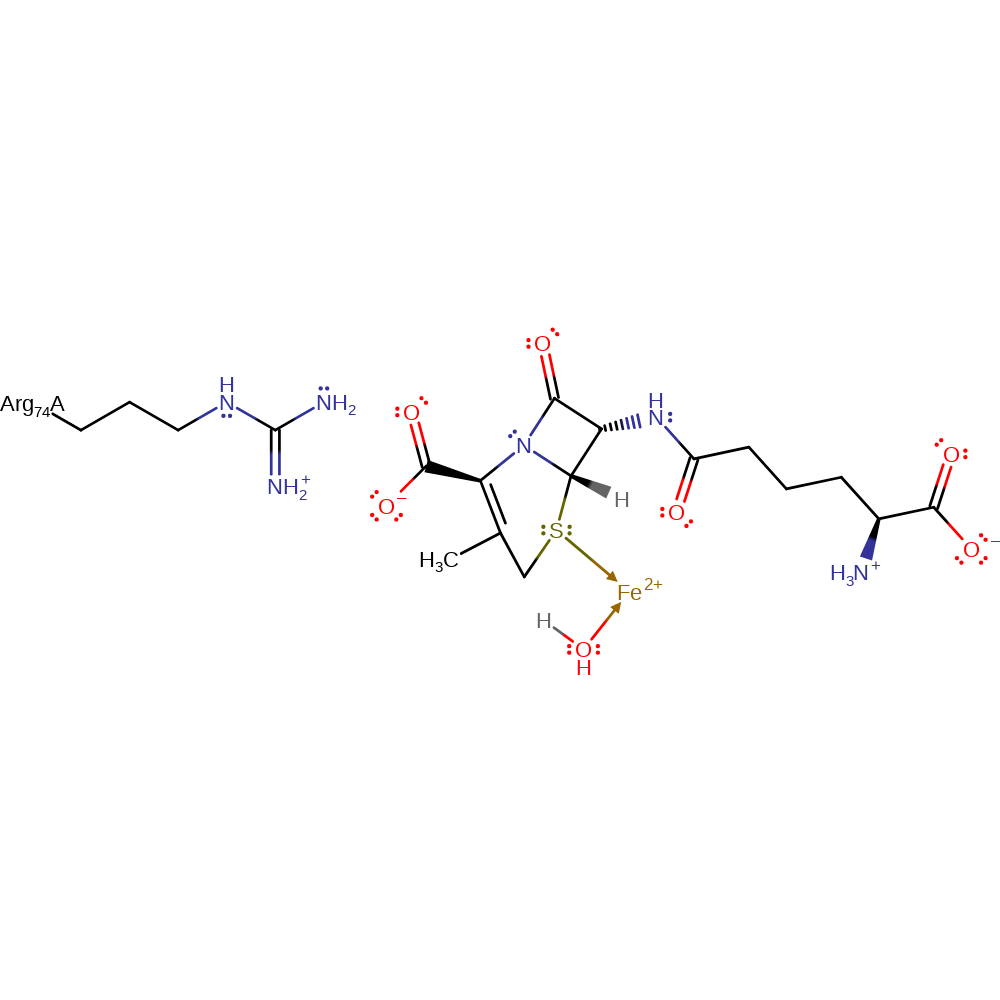 Download:
Download: