Alkanal monooxygenase (FMN-linked)
In the bioluminescence reaction catalysed by bacterial luciferase, a flavin-dependent monooxygenase, reduced riboflavin 5' -phosphate (FMNH2) and a long-chain aliphatic aldehyde are oxidised by molecular oxygen to produce FMN, water, aliphatic carboxylic acid, and visible light with an overall quantum yield of about 0.12. It is intriguing that bacterial luciferase is unique among all known flavin-dependent monooxygenases in catalysing a light-emitting reaction. Vibrio harveyi luciferase is an alpha-beta heterodimer, the proposed earlier to be at a cleft in the alpha subunit.
Reference Protein and Structure
- Sequences
-
P07740
 (1.14.14.3)
(1.14.14.3)
P07739 (1.14.14.3)
(1.14.14.3)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Vibrio harveyi (Bacteria)

- PDB
-
1luc
- BACTERIAL LUCIFERASE
(1.5 Å)



- Catalytic CATH Domains
-
3.20.20.30
 (see all for 1luc)
(see all for 1luc)
Enzyme Reaction (EC:1.14.14.3)
Enzyme Mechanism
Introduction
This proposal represents a modified version of the chemically initiated electron exchange luminescence which predicts that the oxidation potential of the flavin should affect the rate of bioluminescence reaction and found to be so in experiments with substituted FMN analogues [PMID:8422349]. Here the FMN cofactor undergoes a double bond rearrangement that results in the single electron transfer from FMN to dioxygen and a proton transfer from an unidentified base to the dioxygen molecule.Then the FMN and dioxygen radical species undergo a colligation reaction to form the FMN-peroxo adduct. The carbonyl carbon of the aldehyde substrate deprotonates the peroxo-intermediate, which attacks the carbonyl carbon of the aldehyde substrate in a nucleophilic addition. FMN donates a single electron into the peroxo group, causing a homolysis of the O-O bond. His44 deprotonates the intermediate, causing the C-H bond to homolyse, with a single electron being donates to the oxygen radical and one onto the carbon of the cleaved C-H bond. The bound oxygen on the FMN deprotonates His44. The negatively charged oxygen on the intermediate initiates a single electron transfer from the intermediate to the FMN which then emits a photon. The FMN-bound hydroxyl group initiates an intramolecular elimination of water, generating the product FMN.
Catalytic Residues Roles
| UniProt | PDB* (1luc) | ||
| His44 | His44A | Acts as a general base to deprotonate the intermediate, and then protonates the bound oxygen on FMN. | proton acceptor, activator, electrostatic stabiliser, proton donor |
| His45 | His45A | Aids formation of the Flavin 4a-hydroperoxide intermediate. | electrostatic stabiliser, polar/non-polar interaction |
Chemical Components
electron transfer, radical formation, proton transfer, overall reactant used, intermediate formation, colligation, radical termination, bimolecular nucleophilic addition, homolysis, intermediate collapse, radical propagation, intermediate terminated, overall product formed, intramolecular eliminationReferences
- Huang S et al. (1998), Biochemistry, 37, 8614-8614. Identification and Characterization of a Catalytic Base in Bacterial Luciferase by Chemical Rescue of a Dark Mutant. DOI:10.1021/bi985039j. PMID:9622513.
- Romero E et al. (2018), Chem Rev, 118, 1742-1769. Same Substrate, Many Reactions: Oxygen Activation in Flavoenzymes. DOI:10.1021/acs.chemrev.7b00650. PMID:29323892.
- Campbell ZT et al. (2009), Biochemistry, 48, 6085-6094. Crystal structure of the bacterial luciferase/flavin complex provides insight into the function of the beta subunit. DOI:10.1021/bi900003t. PMID:19435287.
- Li CH et al. (2005), Biochemistry, 44, 12970-12977. Active Site Hydrophobicity Is Critical to the Bioluminescence Activity ofVibrio harveyiLuciferase†. DOI:10.1021/bi050935y. PMID:16185065.
- Low JC et al. (2002), Biochemistry, 41, 1724-1731. Functional Roles of Conserved Residues in the Unstructured Loop ofVibrio harveyiBacterial Luciferase†. DOI:10.1021/bi011958p. PMID:11827516.
- Li H et al. (1999), Biochemistry, 38, 4409-4415. Effects of Mutations of the αHis45 Residue ofVibrio harveyiLuciferase on the Yield and Reactivity of the Flavin Peroxide Intermediate†. DOI:10.1021/bi982396l. PMID:10194361.
- Huang S et al. (1997), Biochemistry, 36, 14609-14615. Identification and Characterization of a Catalytic Base in Bacterial Luciferase by Chemical Rescue of a Dark Mutant†. DOI:10.1021/bi9722554. PMID:9402752.
- Eckstein JW et al. (1993), Biochemistry, 32, 404-411. Mechanism of bacterial bioluminescence: 4a,5-Dihydroflavin analogs as models for luciferase hydroperoxide intermediates and the effect of substituents at the 8-position of flavin on luciferase kinetics. DOI:10.1021/bi00053a004. PMID:8422349.

Step 1. FMN undergoes a double bond rearrangement that results in the single electron transfer from FMN to dioxygen and a proton transfer from an unidentified base to the dioxygen molecule.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His45A | electrostatic stabiliser |
| His45A | polar/non-polar interaction |
Chemical Components
electron transfer, radical formation, proton transfer, overall reactant used, intermediate formation
Step 2.
The FMN and dioxygen radical species undergo a colligation reaction to form the FMN-peroxo adduct.
The generation of the excited 4a-hydroxyFMNH emitter originates from the fission of the O-O bond of an intermediate along the reaction mechanism. The intermediate formed in this step can undergo a competing dark decay via the scission of the C4a-O bond.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His45A | electrostatic stabiliser, polar/non-polar interaction |
Chemical Components
colligation, radical termination, intermediate formation
Step 3. The carbonyl carbon of the aldehyde substrate deprotonates the peroxo-intermediate, which attacks the carbonyl carbon of the aldehyde substrate in a nucleophilic addition.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His45A | electrostatic stabiliser, polar/non-polar interaction |
| His44A | activator |
Chemical Components
ingold: bimolecular nucleophilic addition, proton transfer, overall reactant used, intermediate formation
Step 4. FMN donates a single electron into the peroxo group, causing a homolysis of the O-O bond.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His44A | electrostatic stabiliser |
| His45A | electrostatic stabiliser |
Chemical Components
homolysis, electron transfer, radical formation, intermediate collapse, intermediate formation
Step 5. His44 deprotonates the intermediate, causing the C-H bond to homolyse, with a single electron being donates to the oxygen radical and one onto the carbon of the cleaved C-H bond.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His45A | electrostatic stabiliser |
| His44A | proton acceptor |
Chemical Components
proton transfer, radical propagation, intermediate formation
Step 6.
The bound oxygen on the FMN deprotonates His44. The negatively charged oxygen on the intermediate initiates a single electron transfer from the intermediate to the FMN which then emits a photon.
The FMN intermediate formed in this step is the postulated excited state which decays to the flavin-4a-hydroxyde concomitant with emission of light [PMID:8422349].
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His45A | electrostatic stabiliser |
| His44A | proton donor |
Chemical Components
electron transfer, proton transfer, radical termination, intermediate terminated, intermediate formation, overall product formed
Step 7. The FMN-bound hydroxyl group initiates an intramolecular elimination of water, generating the product FMN.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His44A | activator |
Chemical Components
ingold: intramolecular elimination, intermediate collapse, intermediate terminated, overall product formedIntroduction
This mechanism follows the Baeyer-Villiger mechanism which follows that once the FMN-peroxo adduct is formed it can nucleophilically attack the aldehyde substrate forming an intermediate which will on collapse transfer a hydride to the 2nd oxygen of the peroxo adduct resulting in the cleavage of the peroxide bond. This forms the excited state of the C(4a)−hydroxyflavin intermediate, which emits blue-green light to return to the ground-state species. The FMN-bound hydroxyl will initiate an intramolecular elimination and result in the release of water and the production of FMN.
Catalytic Residues Roles
| UniProt | PDB* (1luc) | ||
| Cys106 | Cys106A | Found at the re-face side of the isoalloxazine ring and helps flavin and aldehyde binding. It also stabilises the C4a-peroxyflavin intermediate. | electrostatic stabiliser |
| Asp113 | Asp113A | Located close to the pyrimidine portion on the si-face of the flavin ring and hydrogen bonds to N3 of the isoalloxazine and controls the electrostatic properties near the pyrimidine side of the flavin by forming hydrogen bond networks with neighboring residues. | hydrogen bond acceptor, electrostatic stabiliser |
| Ser227 | Ser227A | Part of the aldehyde bind site and stabilises the negative charge built up on the aldehyde. | hydrogen bond donor, electrostatic stabiliser |
| His44 | His44A | His44 hydrogen bonds to the C4-O stabilising FMN and its intermediate states. Also in other mechanisms is shown to be the catalytic base. | hydrogen bond donor, electrostatic stabiliser |
Chemical Components
electron transfer, homolysis, intermediate formation, intramolecular rearrangement, overall reactant used, bimolecular homolytic addition, colligation, bimolecular nucleophilic addition, unimolecular elimination by the conjugate base, hydride transfer, intermediate collapse, overall product formed, proton transfer, intramolecular elimination, intermediate terminatedReferences
- Romero E et al. (2018), Chem Rev, 118, 1742-1769. Same Substrate, Many Reactions: Oxygen Activation in Flavoenzymes. DOI:10.1021/acs.chemrev.7b00650. PMID:29323892.
- Tinikul R et al. (2016), Adv Biochem Eng Biotechnol, 154, 47-74. Structure, Mechanism, and Mutation of Bacterial Luciferase. DOI:10.1007/10_2014_281. PMID:25487767.
- Campbell ZT et al. (2009), Biochemistry, 48, 6085-6094. Crystal structure of the bacterial luciferase/flavin complex provides insight into the function of the beta subunit. DOI:10.1021/bi900003t. PMID:19435287.
- Huang S et al. (1997), Biochemistry, 36, 14609-14615. Identification and Characterization of a Catalytic Base in Bacterial Luciferase by Chemical Rescue of a Dark Mutant†. DOI:10.1021/bi9722554. PMID:9402752.

Step 1. FMNH2 undergoes a double bond rearrangement which results in the single electron transfer to a dioxygen molecule which results in the homolysis of a bond and production of a superoxide radical.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His44A | electrostatic stabiliser |
| Cys106A | electrostatic stabiliser |
| Asp113A | electrostatic stabiliser |
| Ser227A | electrostatic stabiliser |
| Asp113A | hydrogen bond acceptor |
| His44A | hydrogen bond donor |
Chemical Components
electron transfer, homolysis, intermediate formation, intramolecular rearrangement, overall reactant used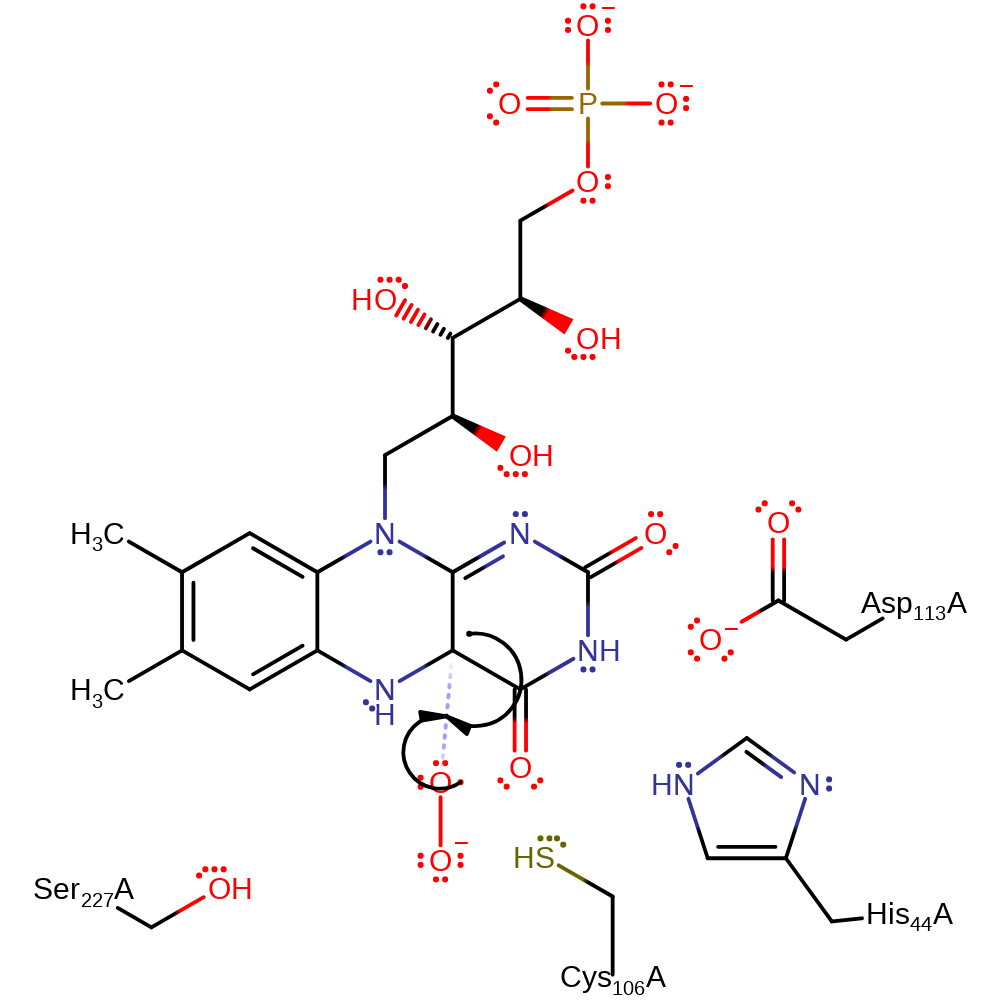
Step 2. The radical species produced colligate to form the FMN-peroxo adduct. The generation of the excited C4a-peroxyflavin emitter originates from the fission of the O-O bond of an intermediate along the reaction mechanism. The intermediate formed in this step can undergo a competing dark decay via the scission of the C4a-O bond.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His44A | electrostatic stabiliser |
| Cys106A | electrostatic stabiliser |
| Asp113A | electrostatic stabiliser |
| Ser227A | electrostatic stabiliser |
| His44A | hydrogen bond donor |
| Asp113A | hydrogen bond acceptor |
Chemical Components
ingold: bimolecular homolytic addition, colligation, intermediate formation
Step 3. The peroxo intermediate nucleophilically attacks the carbonyl carbon of the aldehyde substrate.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His44A | electrostatic stabiliser |
| Cys106A | electrostatic stabiliser |
| Asp113A | electrostatic stabiliser |
| Ser227A | electrostatic stabiliser |
| His44A | hydrogen bond donor |
| Ser227A | hydrogen bond donor |
| Asp113A | hydrogen bond acceptor |
Chemical Components
ingold: bimolecular nucleophilic addition, intermediate formation
Step 4. The intermediate formed undergoes a Baeyer-Villiger reaction which involves the collapse of the oxyanion resulting in the intermediate transfering a hydride to oxygen which results in the cleavage of the peroxide bond. The hydroxyl leaving group bound to FMN will then accept a proton. The intermediate produced here is the excited state which's decay is concomittant with the emission of light.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His44A | electrostatic stabiliser |
| Cys106A | electrostatic stabiliser |
| Asp113A | electrostatic stabiliser |
| Ser227A | electrostatic stabiliser |
| His44A | hydrogen bond donor |
| Ser227A | hydrogen bond donor |
| Asp113A | hydrogen bond acceptor |
Chemical Components
ingold: unimolecular elimination by the conjugate base, hydride transfer, intermediate collapse, intermediate formation, overall product formed, proton transfer
Step 5. The FMN-bound hydroxyl will initiate an intramolecular elimination of water, generating the product FMN.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His44A | electrostatic stabiliser |
| Cys106A | electrostatic stabiliser |
| Asp113A | electrostatic stabiliser |
| Ser227A | electrostatic stabiliser |
| His44A | hydrogen bond donor |
| Ser227A | hydrogen bond donor |
| Asp113A | hydrogen bond acceptor |
Chemical Components
ingold: intramolecular elimination, intermediate collapse, intermediate terminated, overall product formed, proton transferIntroduction
This follows initially a similar reaction as the other proposals where following a double bond rearrangement of FMNH2 and the production if a radical the addition of dioxygen to flavin produces an intermediate which can then nucleophilically attack the aldehyde substrate. The flavin C(4a)-peroxyhemiacetal formed will decomposes producing a dioxirane intermediate and the excited state C(4a)-hydroxyflavin intermediate which will decay to its ground state and emit blue-green light. The intermediate will then undergo an intramolecular elimination which will result in the release of water and produce the final product of FMN.
Catalytic Residues Roles
| UniProt | PDB* (1luc) | ||
| Cys106 | Cys106A | Found at the re-face side of the isoalloxazine ring and helps flavin and aldehyde binding. It also stabilises the C4a-peroxyflavin intermediate. | electrostatic stabiliser |
| Asp113 | Asp113A | Located close to the pyrimidine portion on the si-face of the flavin ring and hydrogen bonds to N3 of the isoalloxazine and controls the electrostatic properties near the pyrimidine side of the flavin by forming hydrogen bond networks with neighbouring residues. | hydrogen bond acceptor, electrostatic stabiliser |
| Ser227 | Ser227A | Part of the aldehyde bind site and stabilises the negative charge built up on the intermediate. | hydrogen bond donor, electrostatic stabiliser |
| His44 | His44A | His44 hydrogen bonds to the C4-O stabilising FMN and its intermediate states. Also in other mechanisms is shown to be the catalytic base. | hydrogen bond donor, electrostatic stabiliser |
Chemical Components
overall reactant used, intramolecular rearrangement, intermediate formation, homolysis, electron transfer, colligation, bimolecular homolytic addition, bimolecular nucleophilic addition, unimolecular elimination by the conjugate base, intermediate collapse, overall product formed, proton transfer, rate-determining step, intramolecular elimination, intermediate terminatedReferences
- Romero E et al. (2018), Chem Rev, 118, 1742-1769. Same Substrate, Many Reactions: Oxygen Activation in Flavoenzymes. DOI:10.1021/acs.chemrev.7b00650. PMID:29323892.
- Tinikul R et al. (2016), Adv Biochem Eng Biotechnol, 154, 47-74. Structure, Mechanism, and Mutation of Bacterial Luciferase. DOI:10.1007/10_2014_281. PMID:25487767.
- Campbell ZT et al. (2009), Biochemistry, 48, 6085-6094. Crystal structure of the bacterial luciferase/flavin complex provides insight into the function of the beta subunit. DOI:10.1021/bi900003t. PMID:19435287.
- Huang S et al. (1997), Biochemistry, 36, 14609-14615. Identification and Characterization of a Catalytic Base in Bacterial Luciferase by Chemical Rescue of a Dark Mutant†. DOI:10.1021/bi9722554. PMID:9402752.

Step 1. FMNH2 undergoes a double bond rearrangement which results in the single electron transfer to a dioxygen molecule which results in the homolysis of a bond and production of a superoxide radical.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His44A | hydrogen bond donor |
| Asp113A | hydrogen bond acceptor |
| Ser227A | electrostatic stabiliser |
| Asp113A | electrostatic stabiliser |
| Cys106A | electrostatic stabiliser |
| His44A | electrostatic stabiliser |
Chemical Components
overall reactant used, intramolecular rearrangement, intermediate formation, homolysis, electron transfer
Step 2. The radical species produced colligate to form the FMN-peroxo adduct. The generation of the excited C4a-peroxyflavin emitter originates from the fission of the O-O bond of an intermediate along the reaction mechanism. The intermediate formed in this step can undergo a competing dark decay via the scission of the C4a-O bond.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp113A | hydrogen bond acceptor |
| His44A | hydrogen bond donor |
| Ser227A | electrostatic stabiliser |
| Asp113A | electrostatic stabiliser |
| Cys106A | electrostatic stabiliser |
| His44A | electrostatic stabiliser |
Chemical Components
intermediate formation, colligation, ingold: bimolecular homolytic addition
Step 3. The peroxo intermediate nucleophilically attacks the carbonyl carbon of the aldehyde substrate.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp113A | hydrogen bond acceptor |
| Ser227A | hydrogen bond donor |
| His44A | hydrogen bond donor |
| Ser227A | electrostatic stabiliser |
| Asp113A | electrostatic stabiliser |
| Cys106A | electrostatic stabiliser |
| His44A | electrostatic stabiliser |
Chemical Components
intermediate formation, ingold: bimolecular nucleophilic addition
Step 4. The flavin C(4a)-peroxyhemiacetal adduct decomposes resulting in the production of dioxirane and the excited state of the C(4a)-hydroxyflavin intermediate which when that decays to ground state will emit blue-green light.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His44A | electrostatic stabiliser |
| Cys106A | electrostatic stabiliser |
| Asp113A | electrostatic stabiliser |
| Ser227A | electrostatic stabiliser |
| His44A | hydrogen bond donor |
| Ser227A | hydrogen bond donor |
| Asp113A | hydrogen bond acceptor |
Chemical Components
ingold: unimolecular elimination by the conjugate base, intermediate collapse, intermediate formation, overall product formed, proton transfer, rate-determining step
Step 5. The FMN-bound hydroxyl group initiates an intramolecular elimination of water, generating the product FMN.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His44A | electrostatic stabiliser |
| Cys106A | electrostatic stabiliser |
| Asp113A | electrostatic stabiliser |
| Ser227A | electrostatic stabiliser |
| His44A | hydrogen bond donor |
| Ser227A | hydrogen bond donor |
| Asp113A | hydrogen bond acceptor |








 Download:
Download: 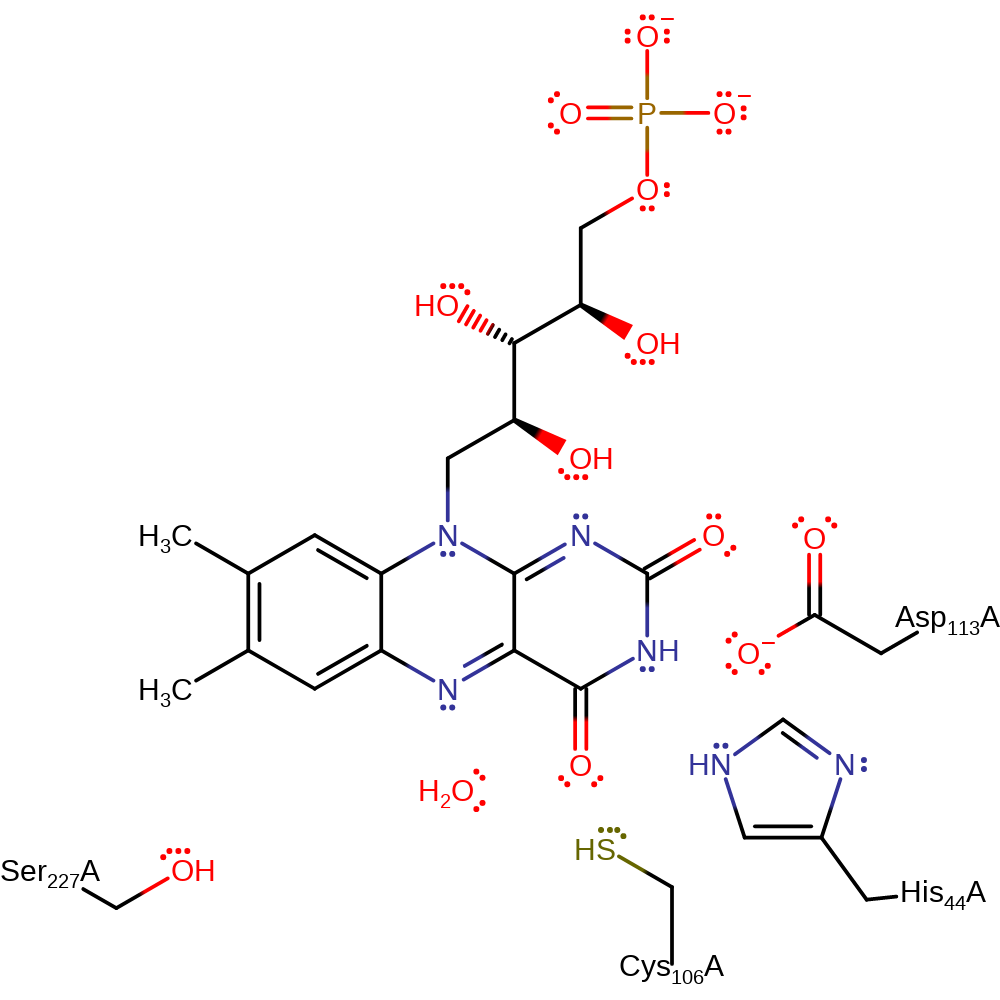 Download:
Download:  Download:
Download: