Ferredoxin hydrogenase
The iron-only hydrogenase from Desulphovibrio desulphuricans is able to catalyse the heterolytic fission of molecular hydrogen to release protons and electrons. It is part of a family of iron-only hydrogenases which all have two iron centres at the active site, in contrast to the better known Ni-Fe hydrogenases with which they share no sequence or structural homology. The mechanism by which the hydrogenases work is of great interest to biologists and engineers alike as it offers the possibility of using hydrogen as a fuel more effectively.
Reference Protein and Structure
- Sequences
-
P07598
 (1.12.7.2)
(1.12.7.2)
P07603 (1.12.7.2)
(1.12.7.2)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Desulfovibrio vulgaris str. Hildenborough (Bacteria)

- PDB
-
1hfe
- 1.6 A RESOLUTION STRUCTURE OF THE FE-ONLY HYDROGENASE FROM DESULFOVIBRIO DESULFURICANS
(1.6 Å)



- Catalytic CATH Domains
-
3.40.50.1780
 3.40.950.10
3.40.950.10  (see all for 1hfe)
(see all for 1hfe)
- Cofactors
- Tetra-mu3-sulfido-tetrairon (3), Iron(2+) (2), Water (4), Iminodimethanethiolate (1) Metal MACiE
Enzyme Reaction (EC:1.12.7.2)
Enzyme Mechanism
Introduction
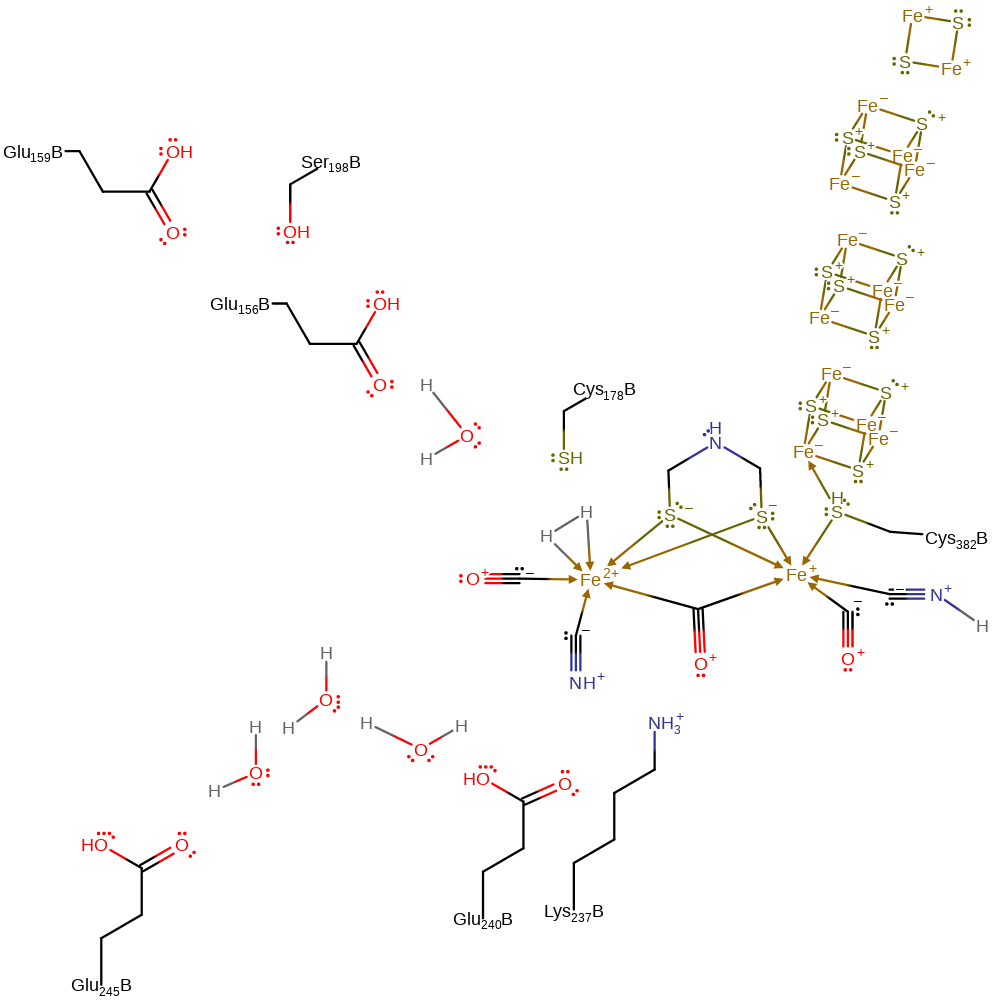
The catalytically active redox state for the binuclear iron centre is, for the formation of hydrogen, Fe(I) Fe(I). This becomes protonated by Lys 237 with the proximal Fe (I) centre accepting a proton. The proximal centre then passes two electrons to the proton to form a hydride ion which stays bonded to the iron, and the distal iron centre assists with this by passing an electron to the proximal centre, forming a Fe (II) Fe (II)H- centre. The bidentate iron ligand DTN is then able to accept a further proton from Cys 178 which allows the hydride ion to act as a base and form molecular hydrogen. This stays bound to the proximal iron centre until an electron is passed from the protein to the centre, at which point H2 is released, generating an Fe (I) Fe (II) centre, which must be further reduced by accepting an electron from the surrounding protein to reform the catalytically active structure. The electrons are presumed to come from the nearby iron sulphur centre, which in turn connects with reducing agents in the cell.
Catalytic Residues Roles
| UniProt | PDB* (1hfe) | ||
| Glu156, Glu159, Cys178, Ser198, Lys237, Glu240, Glu245 | Glu156L(B), Glu159L(B), Cys178L(B), Ser198L(B), Lys237L(B), Glu240L(B), Glu245L(B) | Form one of the proton relay chains that links the active site with bulk solvent. | proton relay, proton acceptor, proton donor |
| Cys382 | Cys382L(B) | Cys382 is a bridging ligand between one of the iron-sulfur clusters and the diiron centre. | metal ligand |
Chemical Components
proton transfer, electron transfer, coordination, heterolysis, substitution (not covered by the Ingold mechanisms), bimolecular nucleophilic substitution, native state of cofactor regenerated, native state of enzyme regeneratedReferences
- Zhou T et al. (2004), Inorg Chem, 43, 923-930. Enzymatic Mechanism of Fe-Only Hydrogenase: Density Functional Study on H−H Making/Breaking at the Diiron Cluster with Concerted Proton and Electron Transfers. DOI:10.1021/ic0342301. PMID:14753812.
- Kubas A et al. (2017), Nat Chem, 9, 88-95. Mechanism of O2 diffusion and reduction in FeFe hydrogenases. DOI:10.1038/nchem.2592. PMID:27995927.
- Sode O et al. (2014), J Chem Phys, 141, 22D527-. Electron transfer activation of a second water channel for proton transport in [FeFe]-hydrogenase. DOI:10.1063/1.4902236. PMID:25494798.
- Myers WK et al. (2014), J Am Chem Soc, 136, 12237-12240. The cyanide ligands of [FeFe] hydrogenase: pulse EPR studies of (13)C and (15)N-labeled H-cluster. DOI:10.1021/ja507046w. PMID:25133957.
- Huynh MT et al. (2014), Inorg Chem, 53, 10301-10311. Computational investigation of [FeFe]-hydrogenase models: characterization of singly and doubly protonated intermediates and mechanistic insights. DOI:10.1021/ic5013523. PMID:25207842.
- Winkler M et al. (2013), Biochim Biophys Acta, 1827, 974-985. Molecular basis of [FeFe]-hydrogenase function: an insight into the complex interplay between protein and catalytic cofactor. DOI:10.1016/j.bbabio.2013.03.004. PMID:23507618.
- Wang N et al. (2013), Dalton Trans, 42, 12059-12071. Reactions of [FeFe]-hydrogenase models involving the formation of hydrides related to proton reduction and hydrogen oxidation. DOI:10.1039/c3dt51371h. PMID:23846321.
- Miyake T et al. (2013), J Biol Inorg Chem, 18, 693-700. Does the environment around the H-cluster allow coordination of the pendant amine to the catalytic iron center in [FeFe] hydrogenases? Answers from theory. DOI:10.1007/s00775-013-1014-4. PMID:23793236.
- Morra S et al. (2012), PLoS One, 7, e48400-. Site saturation mutagenesis demonstrates a central role for cysteine 298 as proton donor to the catalytic site in CaHydA [FeFe]-hydrogenase. DOI:10.1371/journal.pone.0048400. PMID:23133586.
- Knörzer P et al. (2012), J Biol Chem, 287, 1489-1499. Importance of the protein framework for catalytic activity of [FeFe]-hydrogenases. DOI:10.1074/jbc.M111.305797. PMID:22110126.
- Hong G et al. (2011), Biochim Biophys Acta, 1807, 510-517. On understanding proton transfer to the biocatalytic [Fe-Fe](H) sub-cluster in [Fe-Fe]H(2)ases: QM/MM MD simulations. DOI:10.1016/j.bbabio.2011.01.011. PMID:21296047.
- Cornish AJ et al. (2011), J Biol Chem, 286, 38341-38347. Mechanism of proton transfer in [FeFe]-hydrogenase from Clostridium pasteurianum. DOI:10.1074/jbc.M111.254664. PMID:21900241.
- Mulder DW et al. (2011), Structure, 19, 1038-1052. Insights into [FeFe]-hydrogenase structure, mechanism, and maturation. DOI:10.1016/j.str.2011.06.008. PMID:21827941.
- Motiu S et al. (2010), Int J Quantum Chem, 110, 2705-2718. [Fe-Fe]-hydrogenase Reactivated by Residue Mutations as Bridging Carbonyl Rearranges: A QM/MM Study. DOI:10.1002/qua.22381. PMID:26045628.
- Silakov A et al. (2009), Phys Chem Chem Phys, 11, 6592-6599. (14)N HYSCORE investigation of the H-cluster of [FeFe] hydrogenase: evidence for a nitrogen in the dithiol bridge. DOI:10.1039/b905841a. PMID:19639134.
- Dogaru D et al. (2009), Int J Quantum Chem, 110, 1784-1792. Residue Mutations in [Fe-Fe]-hydrogenase Impedes O(2) Binding: A QM/MM Investigation. DOI:10.1002/qua.22331. PMID:20485511.
- Lee JW et al. (2009), Dalton Trans, 8532-8537. Effect of Lewis acid on the structure of a diiron dithiolate complex based on the active site of [FeFe]-hydrogenase assessed by density functional theory. DOI:10.1039/b905786b. PMID:19809728.
- Dogaru D et al. (2009), Int J Quantum Chem, 109, 876-889. Inactivation of [Fe-Fe]-Hydrogenase by O(2). Thermodynamics and Frontier Molecular Orbitals Analyses. DOI:10.1002/qua.21875. PMID:20160838.
- Liu Z et al. (2002), J Chem Phys, 117, 8177-8180. Mechanism of H2 metabolism on Fe-only hydrogenases. DOI:10.1063/1.1519252.
- Liu ZP et al. (2002), J Am Chem Soc, 124, 5175-5182. A Density Functional Theory Study on the Active Center of Fe-Only Hydrogenase: Characterization and Electronic Structure of the Redox States. DOI:10.1021/ja0118690. PMID:11982382.
- Nicolet Y et al. (2002), J Inorg Biochem, 91, 1-8. Fe-only hydrogenases: structure, function and evolution. DOI:10.1016/s0162-0134(02)00392-6. PMID:12121756.
- Nicolet Y et al. (1999), Structure, 7, 13-23. Desulfovibrio desulfuricans iron hydrogenase: the structure shows unusual coordination to an active site Fe binuclear center. DOI:10.1016/s0969-2126(99)80005-7. PMID:10368269.
- Peters JW et al. (1998), Science, 282, 1853-1858. X-ray Crystal Structure of the Fe-Only Hydrogenase (CpI) from Clostridium pasteurianum to 1.8 Angstrom Resolution. DOI:10.1126/science.282.5395.1853. PMID:9836629.
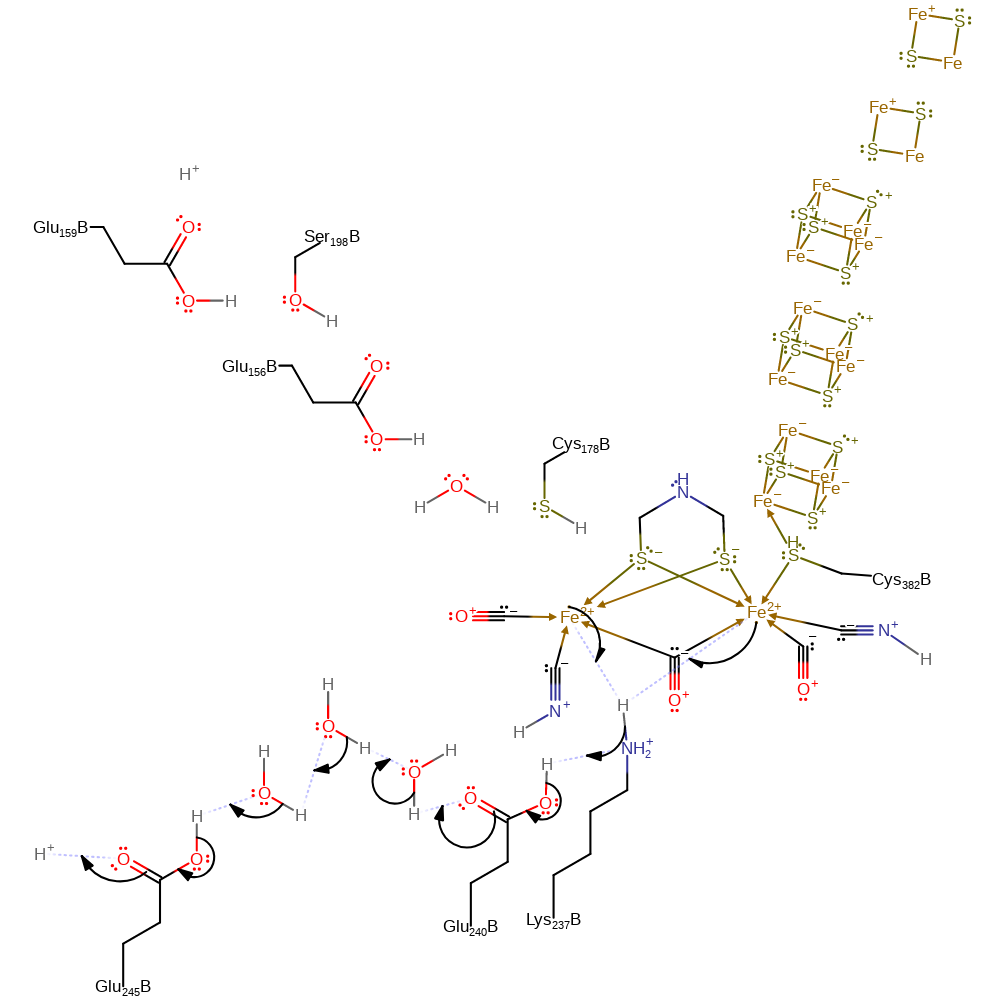
Step 1. Each of the Fe(I) centres donates a single electron to one of the Lys237 protons, forming a 2-electron-3-centre bond bridging the two Fe(II) centres. The loss of the proton from Lys237 initiates a proton relay chain that involves Glu240, three water molecules and Glu245 before terminating in the bulk solvent.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Lys237L(B) | proton donor |
| Cys382L(B) | metal ligand |
| Glu245L(B) | proton donor |
| Lys237L(B) | proton acceptor |
| Glu240L(B) | proton acceptor |
| Glu245L(B) | proton acceptor |
| Glu240L(B) | proton donor |
| Lys237L(B) | proton relay |
| Glu240L(B) | proton relay |
| Glu245L(B) | proton relay |
Chemical Components
proton transfer, electron transfer, coordination, heterolysis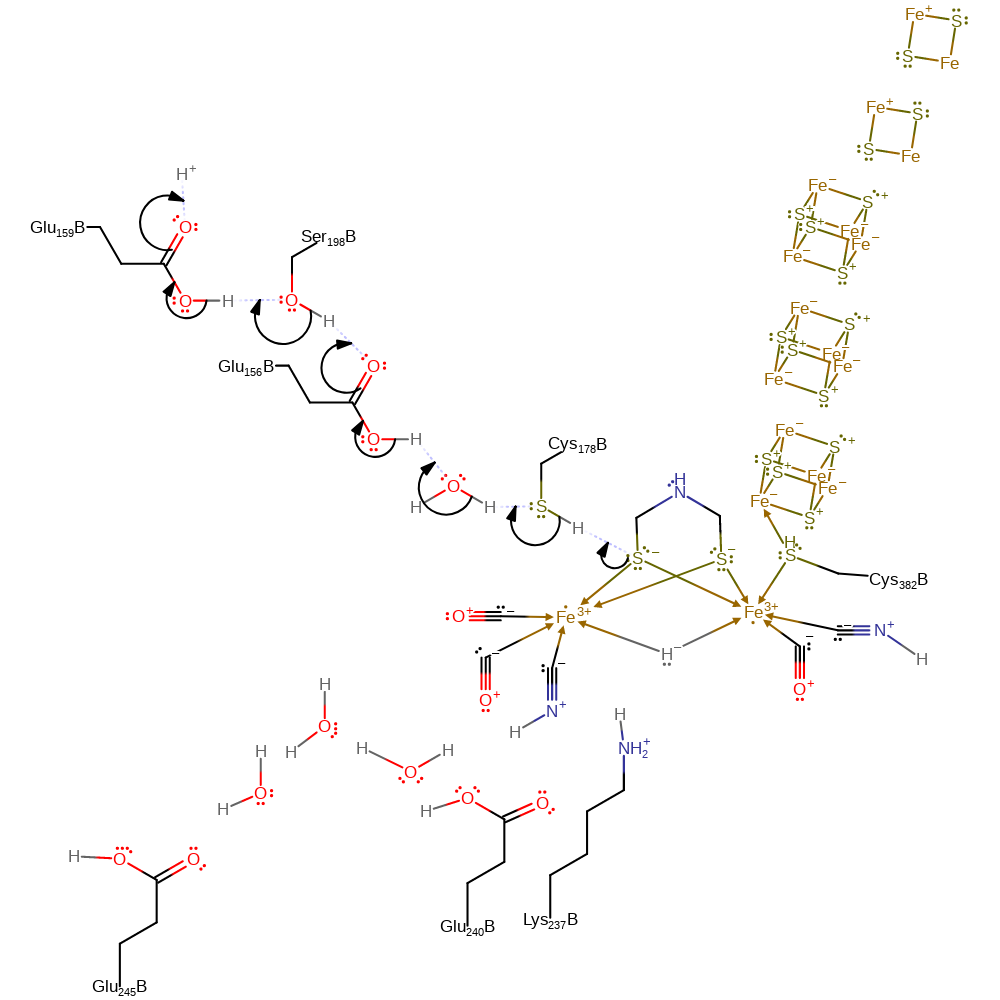
Step 2. One of the iron-bound thiolates deprotonates Cys178, which initiates a proton relay chain that involves water,Glu156, Ser198 and Glu159 before terminating in bulk solvent.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Cys382L(B) | metal ligand |
| Glu159L(B) | proton acceptor |
| Cys178L(B) | proton acceptor |
| Glu156L(B) | proton acceptor |
| Ser198L(B) | proton acceptor |
| Cys178L(B) | proton donor |
| Glu159L(B) | proton donor |
| Ser198L(B) | proton donor |
| Glu156L(B) | proton donor, proton relay |
| Glu159L(B) | proton relay |
| Cys178L(B) | proton relay |
| Ser198L(B) | proton relay |
Chemical Components
proton transfer
Step 3. The bridging hydride deprotonates the thiol, reforming the bound thiolate and a hydrogen molecule bound to one of the Fe(II) centres in a bidentate fashion.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Cys382L(B) | metal ligand |
Chemical Components
proton transfer
Step 4. Ferrocytochrome C2 donates a single electron through three iron-sulfur clusters and to the on the Fe(II) centres. This initiates the coordination of the hydrogen molecule through a single hydrogen atom to both iron centres.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Cys382L(B) | metal ligand |
Chemical Components
substitution (not covered by the Ingold mechanisms), electron transfer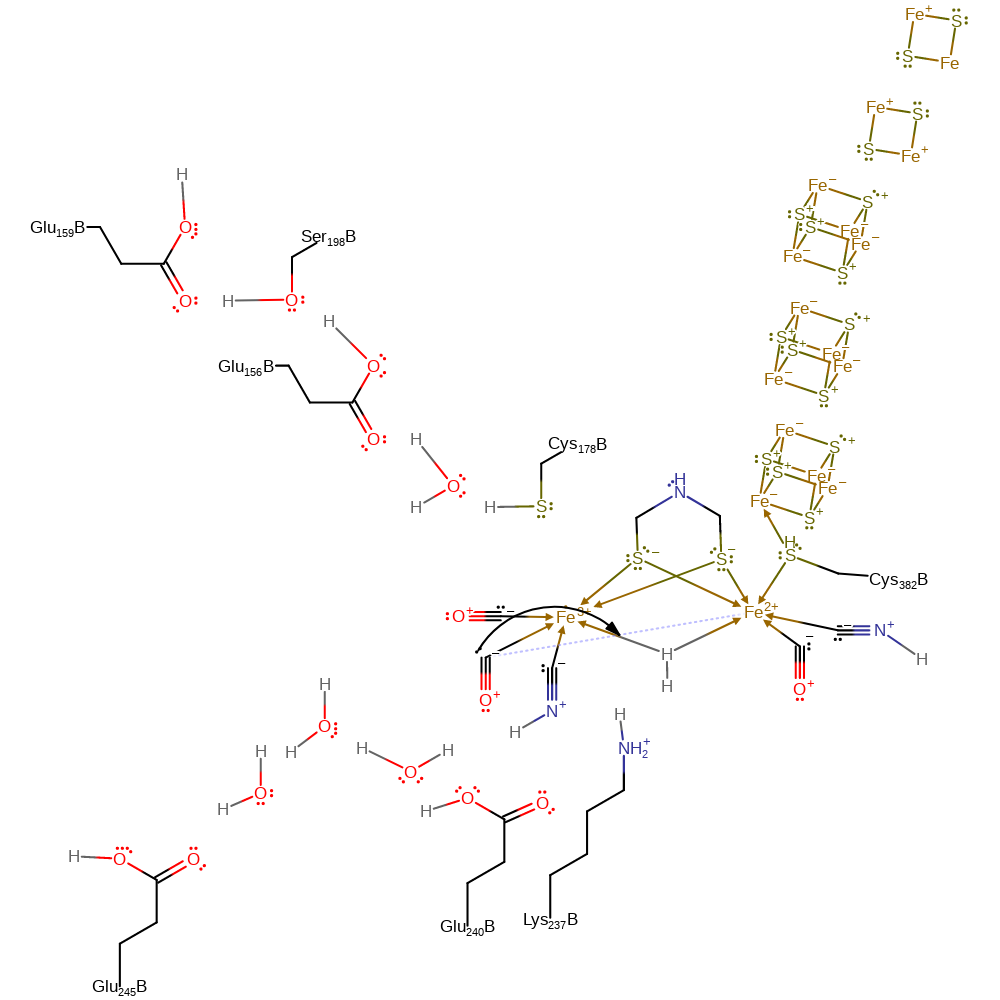
Step 5. One of the carbon monoxide ligands initiates a nucleophilic substitution, resulting in the hydrogen molecule being bound to the Fe(II) centre in a bidentate fashion.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Cys382L(B) | metal ligand |
Chemical Components
ingold: bimolecular nucleophilic substitution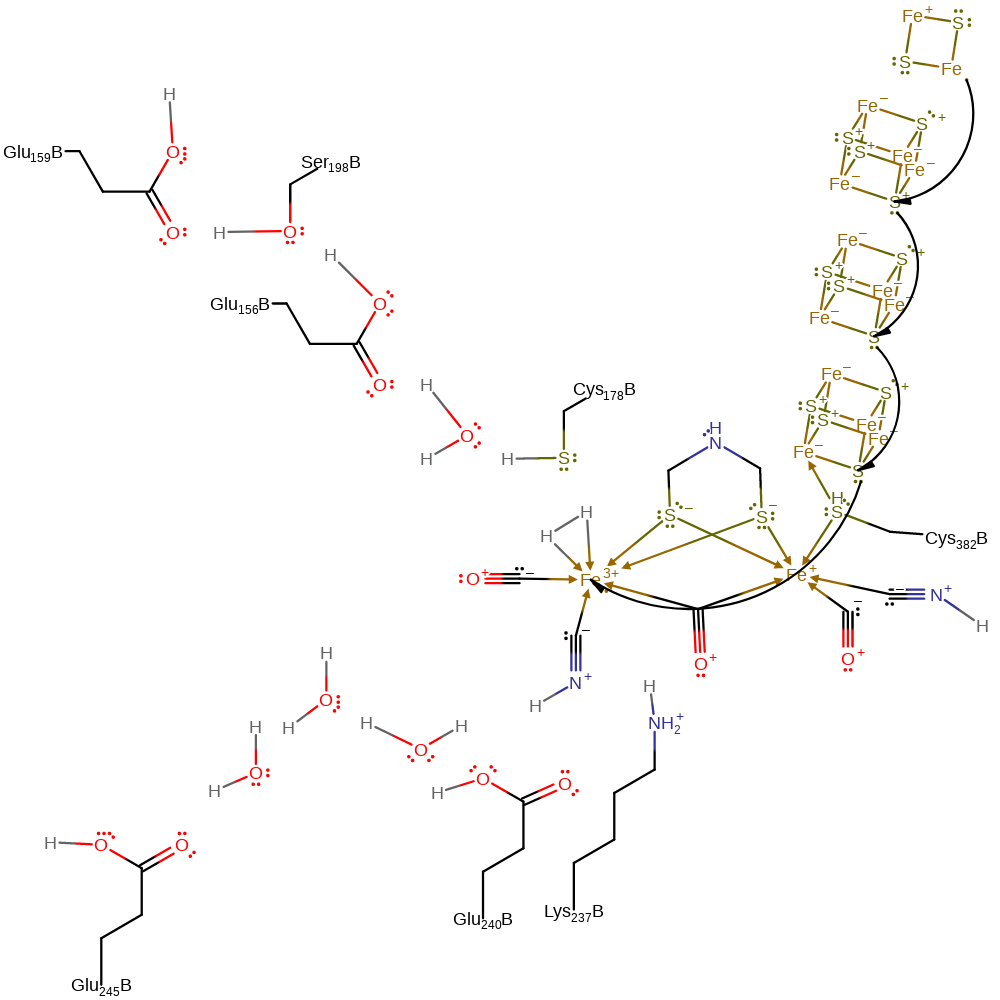
Step 6. Ferrocytochrome C3 donates a second electron through three iron-sulfur clusters to the second Fe(II) centre, initiating the heterolysis of the bond between the hydrogen molecule and Fe(II) centre.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Cys382L(B) | metal ligand |




 Download:
Download: