Deoxyribonuclease IV (phage-T4-induced)
The genetic integrity of cells depends on the concerted action of repair enzymes that recognise and excise damaged bases and mutagenic lesions from DNA. The primary defence against these genotoxic insults is the DNA base excision repair (BER) pathway. The first step of BER is initiated by many distinct DNA glycosylases that each recognise a specific class of damaged DNA nucleotide and cleave the N-C1' glycosidic bond, linking the aberrant base to the deoxyribose sugar. These damage-specific glycosyllases generate as a common product apurinic/apyrimidinic (AP or abasic) sites, which are inherently toxic and mutagenic and thus must be rapidly processed and removed. In the subsequent damage-general steps of single nucleotide BER, an AP endonuclease cleaves the DNA backbone at AP sites, providing a product that is further processed by a DNA deoxyribosephosphodiesterase, a DNA polymerase, and a DNA ligase [PMID:10458614].
Endo IV is an ~30kDa Zn(II) -dependent endonuclease that, unlike the Mg(II) -dependent AP endonuclease III and APE-1, resists inactivation by EDTA. The purified enzyme specifically cleaves the DNA backbone at AP sites and also removes 3'-DNA-blocking groups such as 3' phosphate, 3' phosphoglycolates, and 3' alpha,beta-unsaturated aldehydes that arise from oxidative base damage and the activity of combined glycosylase/lyase enzymes [PMID:10458614].
Reference Protein and Structure
- Sequence
-
P0A6C1
 (3.1.21.2)
(3.1.21.2)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Escherichia coli K-12 (Bacteria)

- PDB
-
1qum
- CRYSTAL STRUCTURE OF ESCHERICHIA COLI ENDONUCLEASE IV IN COMPLEX WITH DAMAGED DNA
(1.55 Å)



- Catalytic CATH Domains
-
3.20.20.150
 (see all for 1qum)
(see all for 1qum)
- Cofactors
- Zinc(2+) (3) Metal MACiE
Enzyme Reaction (EC:3.1.21.2)
Enzyme Mechanism
Introduction
Hydrolysis proceeds through a pentacoordinate transition state where the unesterified phosphate oxygen that bridges Zn2 and Zn3 remains bound to its cognate metal ions. Initial binding of Endo IV to an extrahelical AP site is constrained by the intact target P-O3' covalent bond, and in the pretransition state, the bridging hydroxide between Zn1 and Zn2 would be positioned ideally for an in-line attack on the phosphate. Glu-261, which is also a Zn2 ligand, may assist in orienting and activating the attacking nucleophile. Charge neutralisation of the phosphate group by interaction with the three Zn2+ ions likely renders the phosphorus atom susceptible to nucleophilic substitution, and the pentacoordinate transition state resulting from attack by the bridging hydroxide is stabilised by all the three metal ions. As this transition state collapses to the reaction products, the stereochemical configuration of the phosphate is inverted and the developing negative charge at O3' stabilised by interactions with Zn3 [PMID:10458614].
Catalytic Residues Roles
| UniProt | PDB* (1qum) | ||
| Arg37, Tyr72 | Arg37A(D), Tyr72A(D) | Stabilises the pentavalent negatively charged transition state. | transition state stabiliser |
| His109, Glu145, Asp179, His216, Glu261, His231, His182, Asp229, His69 | His109A(D), Glu145A(D), Asp179A(D), His216A(D), Glu261A(D), His231A(D), His182A(D), Asp229A(D), His69A(D) | Binds one of the Zn(II) ions. | metal ligand |
Chemical Components
bimolecular nucleophilic substitution, overall reactant used, overall product formed, rate-determining step, proton transfer, inferred reaction step, native state of enzyme regenerated, intermediate terminatedReferences
- Hosfield DJ et al. (1999), Cell, 98, 397-408. Structure of the DNA repair enzyme endonuclease IV and its DNA complex: double-nucleotide flipping at abasic sites and three-metal-ion catalysis. PMID:10458614.
- Garcin ED et al. (2008), Nat Struct Mol Biol, 15, 515-522. DNA apurinic-apyrimidinic site binding and excision by endonuclease IV. DOI:10.1038/nsmb.1414. PMID:18408731.
- Ivanov I et al. (2007), Proc Natl Acad Sci U S A, 104, 1465-1470. Unraveling the three-metal-ion catalytic mechanism of the DNA repair enzyme endonuclease IV. DOI:10.1073/pnas.0603468104. PMID:17242363.
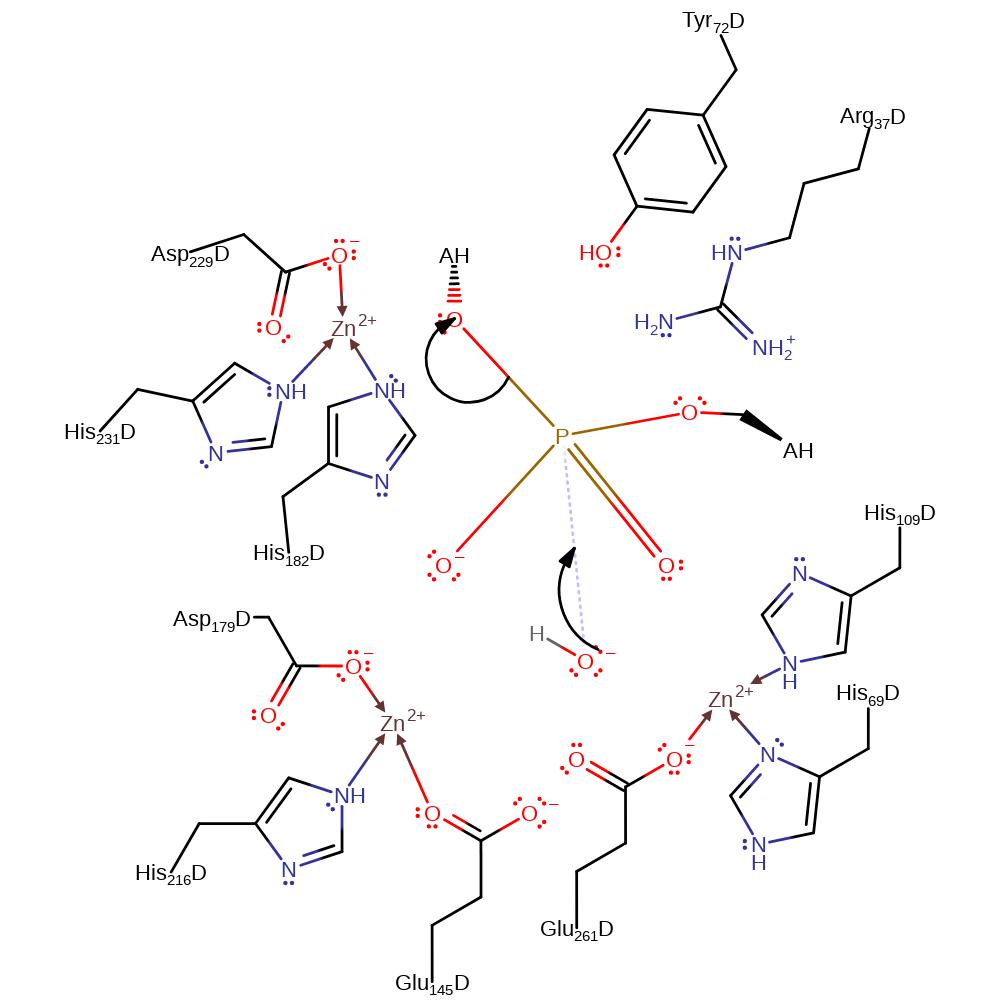
Step 1. Zinc activated water initiates a nucleophilic attack on the DNA phosphate, concomitantly eliminating the 5'-ribose in a substitution reaction which proceeds through a pentavalent transition state.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp229A(D) | metal ligand |
| His231A(D) | metal ligand |
| His182A(D) | metal ligand |
| Asp179A(D) | metal ligand |
| His216A(D) | metal ligand |
| Glu145A(D) | metal ligand |
| Glu261A(D) | metal ligand |
| His109A(D) | metal ligand |
| His69A(D) | metal ligand |
| Tyr72A(D) | transition state stabiliser |
| Arg37A(D) | transition state stabiliser |
Chemical Components
ingold: bimolecular nucleophilic substitution, overall reactant used, overall product formed, rate-determining step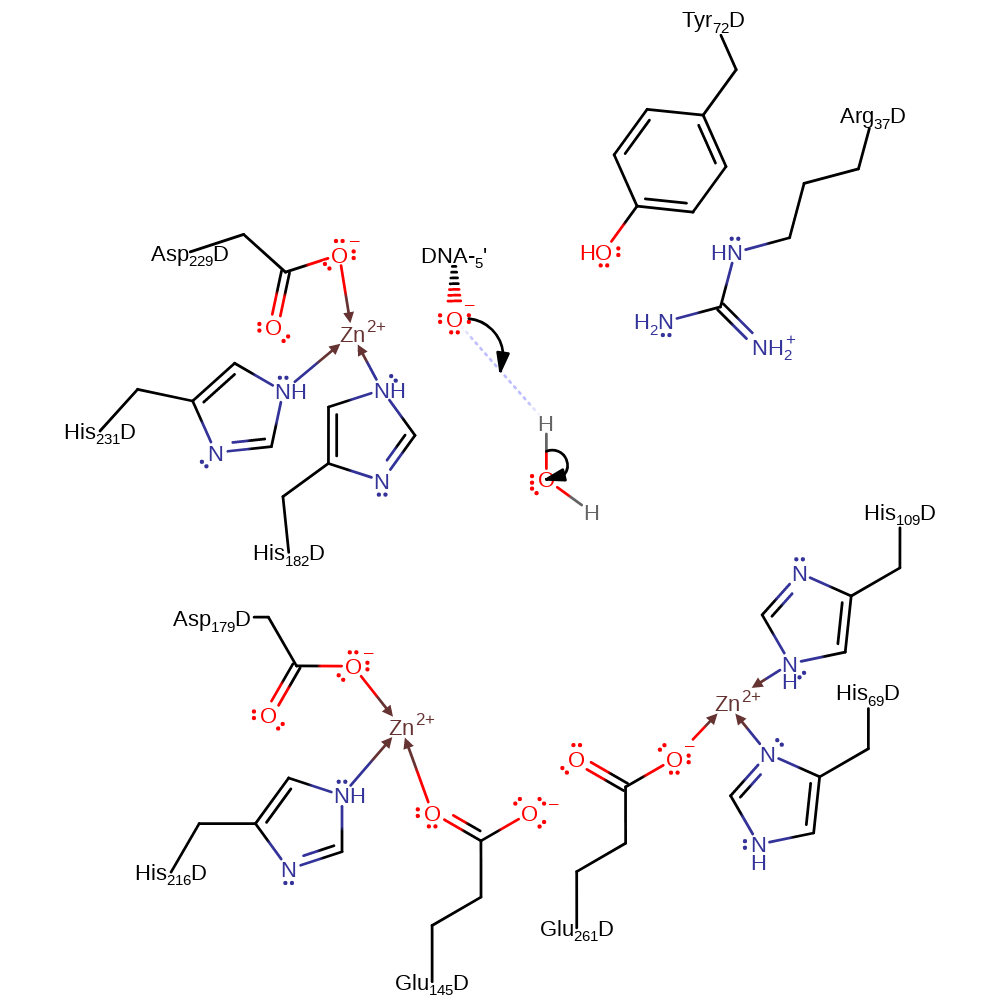
Step 2. The DNA 3'-hydroxyl deprotonates a water molecule to regenerate the active site and the 3'hydroxyl product in an inferred return step.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp229A(D) | metal ligand |
| His231A(D) | metal ligand |
| His182A(D) | metal ligand |
| Asp179A(D) | metal ligand |
| Glu261A(D) | metal ligand |
| Glu145A(D) | metal ligand |
| His69A(D) | metal ligand |
| His109A(D) | metal ligand |
| His216A(D) | metal ligand |
| Arg37A(D) | transition state stabiliser |
| Tyr72A(D) | transition state stabiliser |




 Download:
Download: