Adenylate cyclase
Adenylate cyclase converts adenosine triphosphate (ATP) into cyclic adenosine monophosphate (cAMP), a ubiquitous second messenger that regulates many functions. Many signalling pathways triggered by hormones and other biological processes converge on and regulate the integral membrane enzyme, including those of the heterotrimeric GTP-binding proteins. There are over 30 related proteins, including bacterial and fungal adenylyl cyclases as well as guanylyl cyclases.
Reference Protein and Structure
- Sequences
-
P26769
 (4.6.1.1)
(4.6.1.1)
P30803 (4.6.1.1)
(4.6.1.1)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Rattus norvegicus (Norway rat)

- PDB
-
1cju
- COMPLEX OF GS-ALPHA WITH THE CATALYTIC DOMAINS OF MAMMALIAN ADENYLYL CYCLASE: COMPLEX WITH BETA-L-2',3'-DIDEOXYATP AND MG
(2.8 Å)



- Catalytic CATH Domains
-
3.30.70.1230
 (see all for 1cju)
(see all for 1cju)
- Cofactors
- Magnesium(2+) (2) Metal MACiE
Enzyme Reaction (EC:4.6.1.1)
Enzyme Mechanism
Introduction
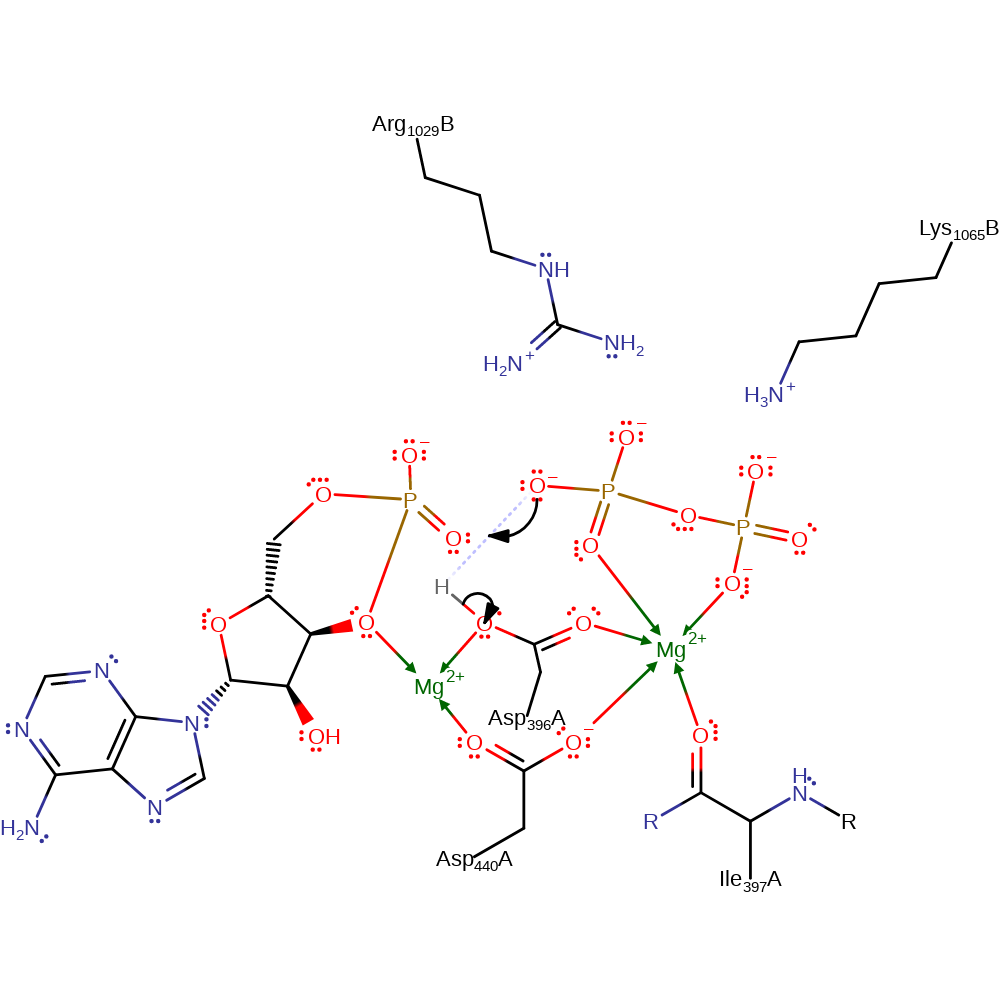
The alpha-phosphate deprotonates the 3'-hydroxyl of ATP, which in turn attacks the phosphorus atom in a nucleophilic addition, forming a cyclic pentacoordinate phosphorus intermediate. The cyclic pentacoordinate phosphorus intermediate collapses eliminating the diphosphate molecule. One or two magnesium ions have been suggested as playing a part in the catalytic mechanism, for example by stabilising the transition state, but they are not present in the PDB file.
Catalytic Residues Roles
| UniProt | PDB* (1cju) | ||
| Asp521 | Asp440(77)A | Forms part of the Magnesium 1 binding site. | metal ligand |
| Arg1029 | Arg1029(160)B | Stabilises negatively charged transition state. Ensures that the transition state for the concerted phosphoryl transfer is "tight". | electrostatic stabiliser |
| Asp477, Asp521, Lys1065 | Asp396(33)A, Asp440(77)A, Lys1065(196)B | Facilitate proton transfer from the ribosyl 3′O to the γ phosphate O2-gamma by decreasing the difference in the pKa values of the proton donor and acceptor and by anchoring the metal ions and hence reducing the free energy barrier for the proton transfer. | metal ligand, proton acceptor, proton donor |
| Ile478 (main-C) | Ile397(34)A (main-C) | Forms part of the Magnesium 2 binding site. | metal ligand |
Chemical Components
cyclisation, proton transfer, intramolecular eliminationReferences
- Tesmer JJ et al. (1997), Science, 278, 1907-1916. Crystal Structure of the Catalytic Domains of Adenylyl Cyclase in a Complex with Gs·GTPS. DOI:10.1126/science.278.5345.1907. PMID:9417641.
- Hahn DK et al. (2015), Biochemistry, 54, 6252-6262. Catalytic Mechanism of Mammalian Adenylyl Cyclase: A Computational Investigation. DOI:10.1021/acs.biochem.5b00655. PMID:26393535.
- Lee Y et al. (2004), J Phys Chem B, 108, 4508-4515. Structure and Reaction in the Active Site of Mammalian Adenylyl Cyclase. DOI:10.1021/jp036564h.
- Serfass L et al. (2001), Arch Biochem Biophys, 387, 47-56. Calcium Ion Downregulates Soluble Guanylyl Cyclase Activity: Evidence for a Two-metal Ion Catalytic Mechanism. DOI:10.1006/abbi.2000.2090. PMID:11368183.
- Tesmer JJ et al. (2000), Biochemistry, 39, 14464-14471. Molecular basis for P-site inhibition of adenylyl cyclase. PMID:11087399.
- Tesmer JJ et al. (1999), Science, 285, 756-760. Two-Metal-Ion Catalysis in Adenylyl Cyclase. DOI:10.1126/science.285.5428.756. PMID:10427002.
- Hurley JH (1999), J Biol Chem, 274, 7599-7602. Structure, Mechanism, and Regulation of Mammalian Adenylyl Cyclase. DOI:10.1074/jbc.274.12.7599.
- Yan SZ et al. (1997), J Biol Chem, 272, 12342-12349. The Conserved Asparagine and Arginine Are Essential for Catalysis of Mammalian Adenylyl Cyclase. DOI:10.1074/jbc.272.19.12342. PMID:9139678.
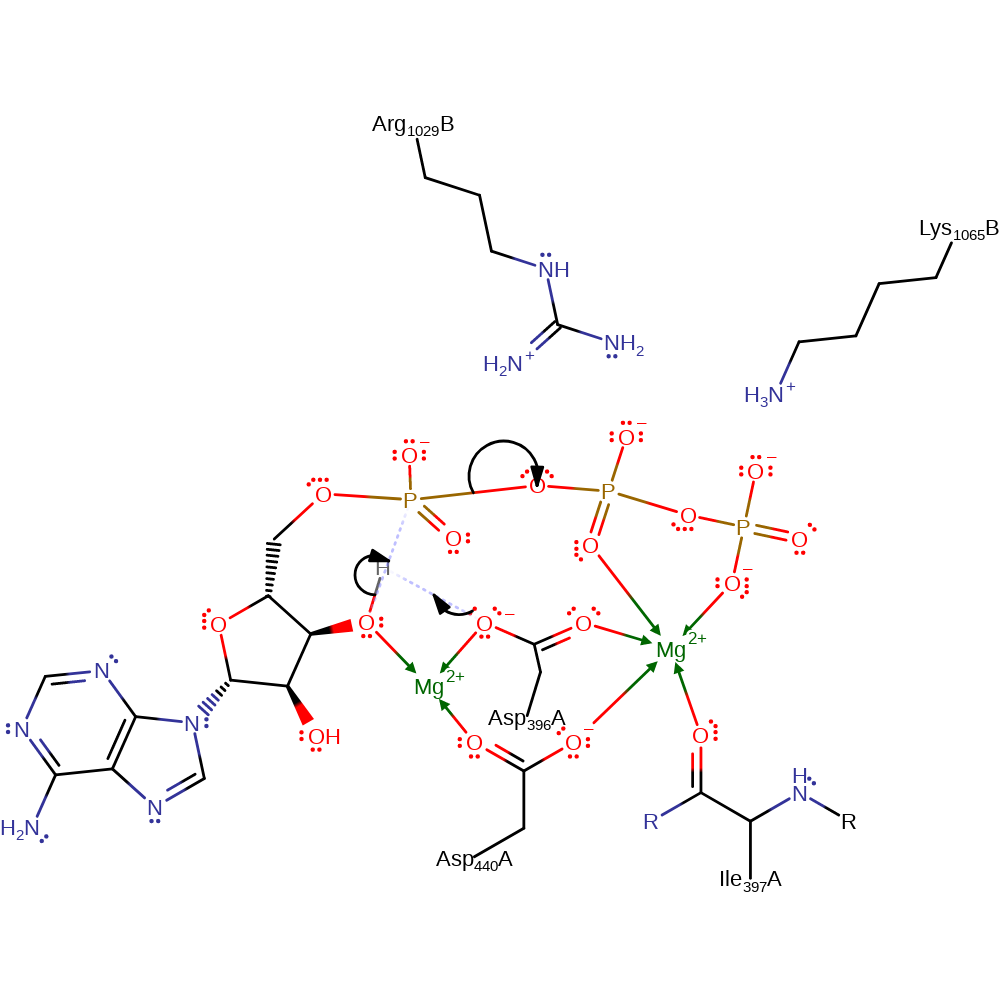
Step 1. Asp396 deprotonates the 3'-hydroxyl of ATP, which in turn attacks the phosphorus atom in a nucleophilic addition, forming a cyclic pentacoordinate phosphorus intermediate.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp440(77)A | metal ligand |
| Asp396(33)A | metal ligand |
| Ile397(34)A (main-C) | metal ligand |
| Arg1029(160)B | electrostatic stabiliser |
| Lys1065(196)B | electrostatic stabiliser |
| Asp396(33)A | proton acceptor |
Chemical Components
cyclisation, proton transfer, ingold: intramolecular eliminationCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp440(77)A | metal ligand |
| Asp396(33)A | metal ligand |
| Ile397(34)A (main-C) | metal ligand |
| Asp396(33)A | proton donor |



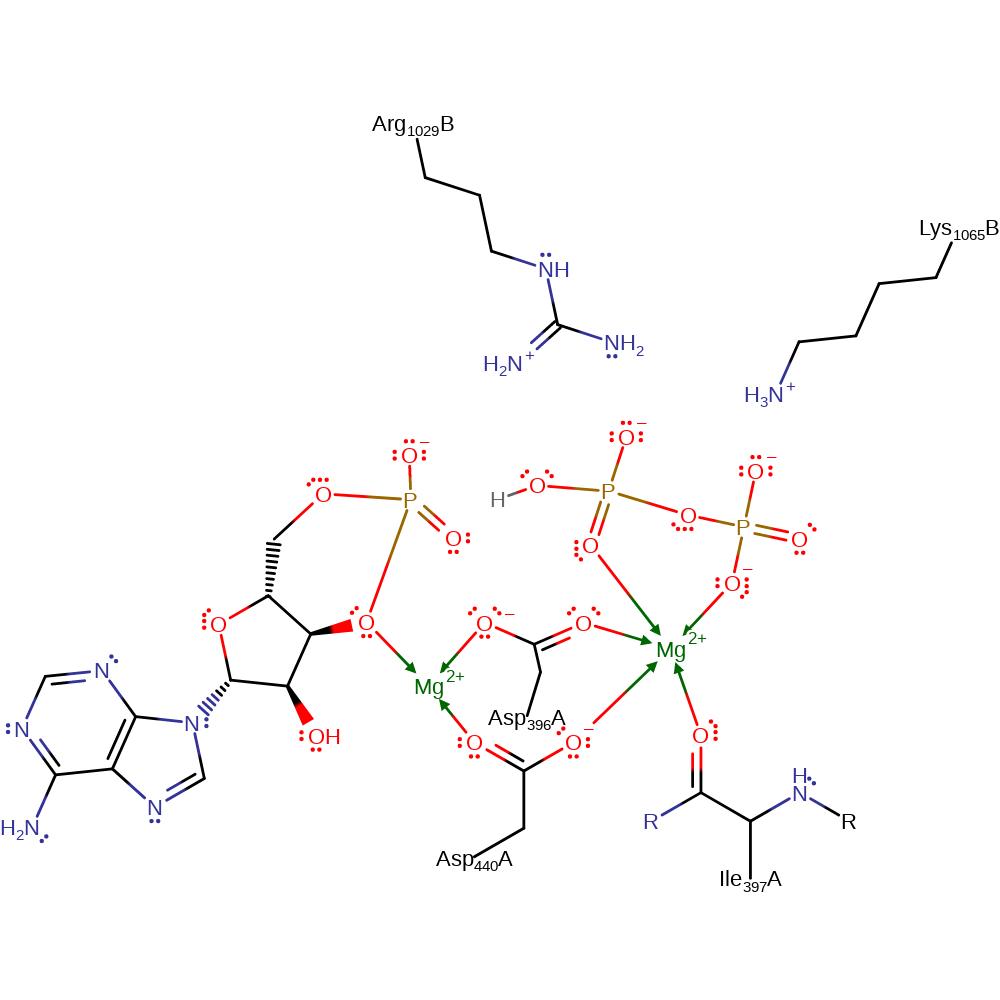 Download:
Download: