4-chlorobenzoyl-CoA dehalogenase
Chlorobenzoate dehalogenase catalyses the hydrolysis of chlorobenzoyl-CoA to hydroxybenzoyl-CoA. This reaction is used by bacteria as part of a three enzyme pathway for the utilisation of chlorinated organic compounds as a carbons source. The chlorobenzoate dehalogenase step is the second in the pathway and is structurally related to the crotonase-like superfamily of enzymes found in the beta-oxidation cycle.
Since chlorinated organic compounds are thought to have only been present in the biosphere in significant amounts for the latter half of this century due to industrial production. The potential for bacteria to evolve new degradation pathways within decades of exposure to a new compound offers the possibility of bioremediation of environmentally hazardous and toxic substances.
Reference Protein and Structure
- Sequence
-
A5JTM5
 (3.8.1.7)
(3.8.1.7)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Pseudomonas sp. CBS3 (Bacteria)

- PDB
-
1nzy
- 4-CHLOROBENZOYL COENZYME A DEHALOGENASE FROM PSEUDOMONAS SP. STRAIN CBS-3
(1.8 Å)



- Catalytic CATH Domains
-
3.90.226.10
 (see all for 1nzy)
(see all for 1nzy)
Enzyme Mechanism
Introduction
Aspartate 145 attacks the chlorobenzoate ring at the chlorinated carbon forming an arylated enzyme intermediate (Meisenheimer intermediate). The chloride then leaves and the enzyme returns to the resting state by attack of an activated water on the acyl carbon. Histidine 90 is thought to be the general base which activates the water based on mutagenic studies. The backbone amides of phenylalanine 64 and glycine 114 form an oxyanion hole for the stabilisation of the tetrahedral intermediate formed in the hydrolytic step [PMID:8679561].
Catalytic Residues Roles
| UniProt | PDB* (1nzy) | ||
| Ala86 (main-C) | Ala86A (main-C) | The main chain carbonyl activates the His90 for its general acid/base function. | activator, hydrogen bond acceptor |
| His90 | His90A | General acid/base that abstracts a proton from Asp145, and later by the nucleophilic water. It is returned to its initial protonation state both times by the leaving group. | hydrogen bond acceptor, hydrogen bond donor, proton acceptor, proton donor |
| Gly114 (main-N), Phe64 (main-N) | Gly114A (main-N), Phe64A (main-N) | Back bond amide forms the oxyanion hole. | hydrogen bond donor, electrostatic stabiliser |
| Trp137 | Trp137A | Stabilises the reaction intermediates. | hydrogen bond donor, electrostatic stabiliser |
| Asp145 | Asp145A | Acts as the catalytic nucleophile. | covalently attached, hydrogen bond acceptor, hydrogen bond donor, nucleophile, proton acceptor, proton donor, electrofuge, electrophile |
Chemical Components
aromatic bimolecular nucleophilic addition, overall reactant used, enzyme-substrate complex formation, intermediate formation, proton transfer, aromatic unimolecular elimination by the conjugate base, overall product formed, enzyme-substrate complex cleavage, intermediate collapse, bimolecular nucleophilic addition, unimolecular elimination by the conjugate base, native state of enzyme regenerated, intermediate terminatedReferences
- Benning MM et al. (1996), Biochemistry, 35, 8103-8109. Structure of 4-Chlorobenzoyl Coenzyme A Dehalogenase Determined to 1.8 Å Resolution: An Enzyme Catalyst Generated via Adaptive Mutation†,‡. DOI:10.1021/bi960768p. PMID:8679561.
- Xie D et al. (2005), J Phys Chem B, 109, 5259-5266. Theoretical Study of General Base-Catalyzed Hydrolysis of Aryl Esters and Implications for Enzymatic Reactions. DOI:10.1021/jp0506181. PMID:16863192.
- Xu D et al. (2005), FEBS Lett, 579, 4249-4253. Electrostatic influence of active-site waters on the nucleophilic aromatic substitution catalyzed by 4-chlorobenzoyl-CoA dehalogenase. DOI:10.1016/j.febslet.2005.06.056. PMID:16051230.
- Xu D et al. (2004), Chem Commun (Camb), 892-. A QM/MM study of a nucleophilic aromatic substitution reaction catalyzed by 4-chlorobenzoyl-CoA dehalogenase. DOI:10.1039/b401159g. PMID:15045116.
- Zheng Y et al. (1997), J Am Chem Soc, 119, 3868-3877. On the Dehalogenation Mechanism of 4-Chlorobenzoyl CoA by 4-Chlorobenzoyl CoA Dehalogenase: Insights from Study Based on the Nonenzymatic Reaction. DOI:10.1021/ja970114j.
- Clarkson J et al. (1997), Biochemistry, 36, 10192-10199. Raman Study of the Polarizing Forces Promoting Catalysis in 4-Chlorobenzoate-CoA Dehalogenase†. DOI:10.1021/bi970941x. PMID:9254617.
- Yang G et al. (1996), Biochemistry, 35, 10879-10885. Identification of Active Site Residues Essential to 4-Chlorobenzoyl−Coenzyme A Dehalogenase Catalysis by Chemical Modification and Site Directed Mutagenesis†. DOI:10.1021/bi9609533. PMID:8718880.

Step 1. His90 deprotonates Asp145, which initiates a nucleophilic attack on the C4 of the 4-chlorobenzoyl-coenzyme A in an addition reaction. The conjugated double bonds rearrange to form an oxyanion, which is stabilised by the main chain amides of Phe64 and Gly114.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Gly114A (main-N) | hydrogen bond donor |
| Trp137A | hydrogen bond donor |
| Ala86A (main-C) | hydrogen bond acceptor, activator |
| Asp145A | hydrogen bond donor, hydrogen bond acceptor |
| His90A | hydrogen bond acceptor, hydrogen bond donor |
| Phe64A (main-N) | hydrogen bond donor |
| Asp145A | proton donor |
| His90A | proton acceptor |
| Asp145A | nucleophile |
Chemical Components
ingold: aromatic bimolecular nucleophilic addition, overall reactant used, enzyme-substrate complex formation, intermediate formation, proton transfer
Step 2. The oxyanion collapses with rearrangement of the conjugated double bonds, eliminating chlorine, which deprotonates the His90.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Gly114A (main-N) | hydrogen bond donor, electrostatic stabiliser |
| Trp137A | hydrogen bond donor |
| Ala86A (main-C) | hydrogen bond acceptor |
| Asp145A | covalently attached, hydrogen bond acceptor |
| His90A | hydrogen bond donor |
| Phe64A (main-N) | hydrogen bond donor, electrostatic stabiliser |
| His90A | proton donor |
Chemical Components
ingold: aromatic unimolecular elimination by the conjugate base, overall product formed, enzyme-substrate complex cleavage, intermediate collapse, intermediate formation, proton transfer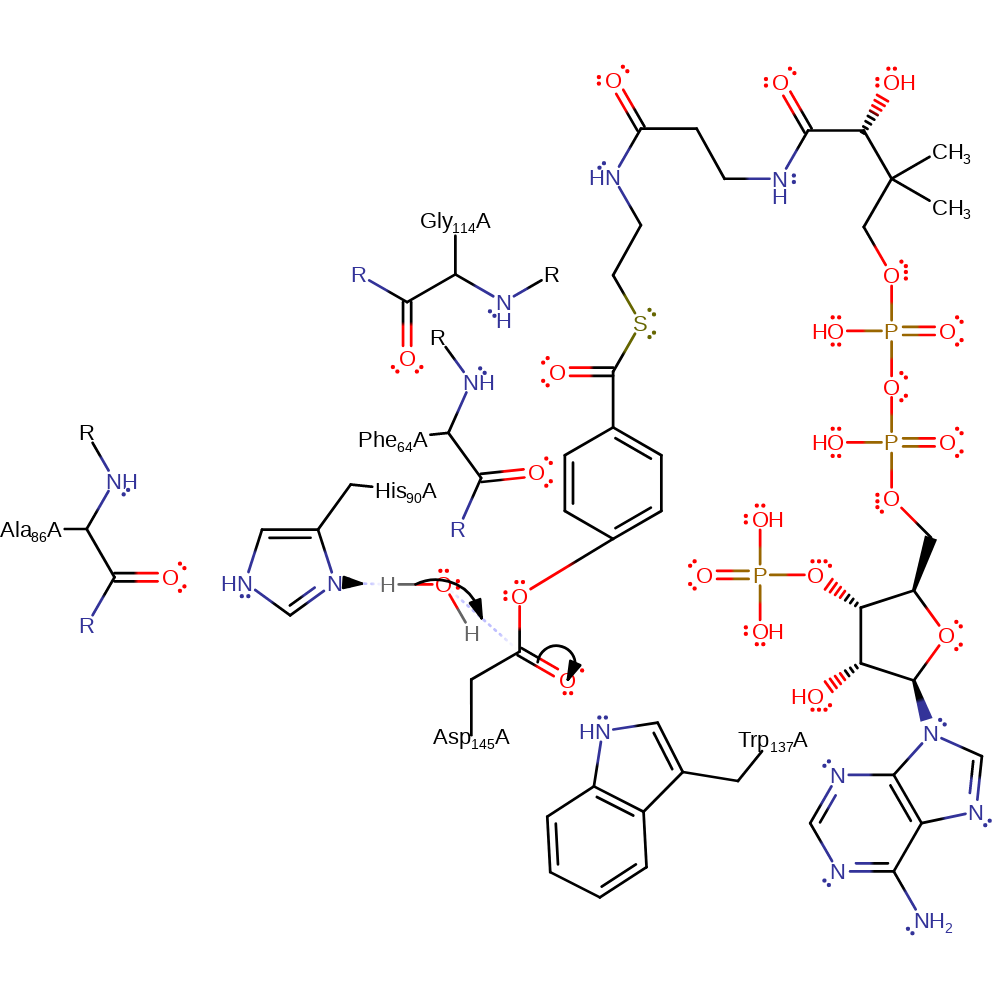
Step 3. His90 deprotonates water, which initiates a nucleophilic attack on the carboxylic carbon of the covalently attached Asp145, forming a new oxyanion, which is stabilised by Trp137.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Gly114A (main-N) | hydrogen bond donor |
| Trp137A | hydrogen bond donor |
| Ala86A (main-C) | hydrogen bond acceptor, activator |
| Asp145A | covalently attached, hydrogen bond acceptor |
| His90A | hydrogen bond acceptor, hydrogen bond donor |
| Phe64A (main-N) | hydrogen bond donor |
| His90A | proton acceptor |
| Asp145A | electrophile |
Chemical Components
ingold: bimolecular nucleophilic addition, enzyme-substrate complex formation, intermediate formation, proton transfer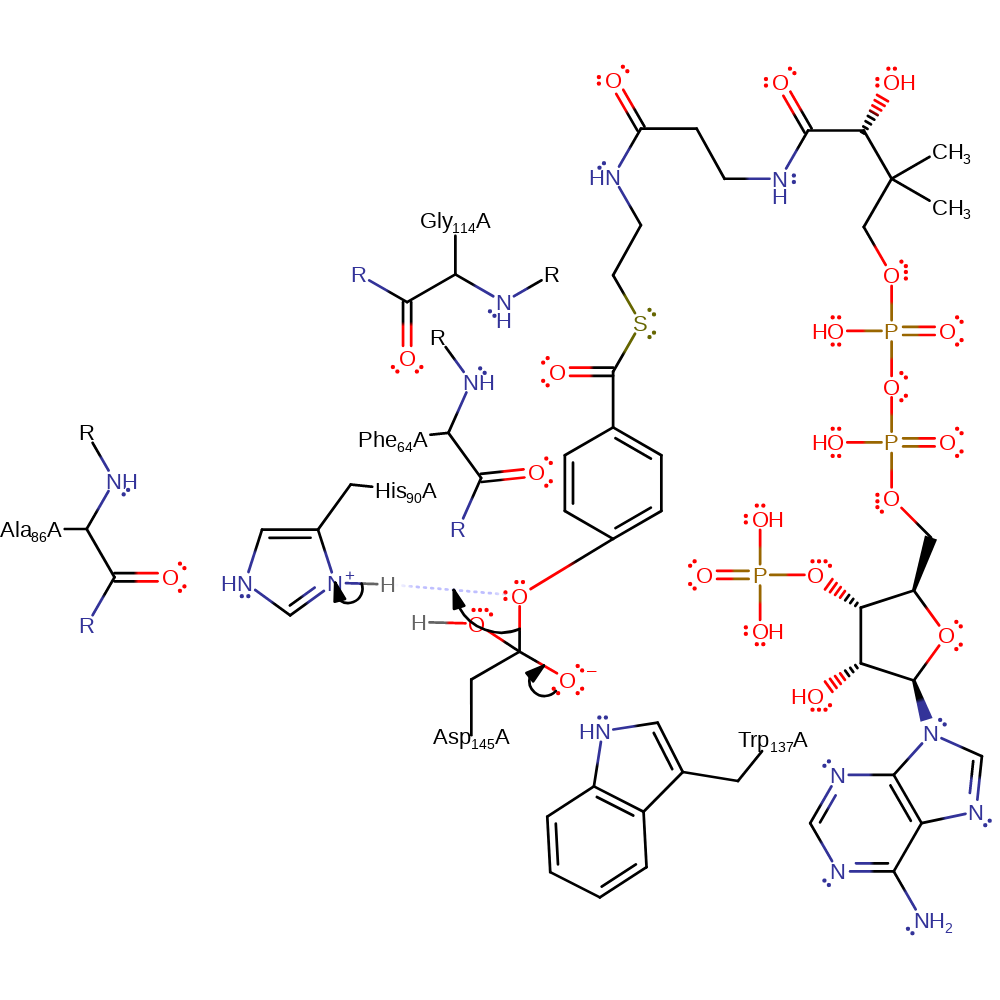
Step 4. The oxyanion collapses, eliminating Asp145 as an electrofuge. The newly formed phenolic oxygen deprotonates His90.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Gly114A (main-N) | hydrogen bond donor |
| Trp137A | hydrogen bond donor, electrostatic stabiliser |
| Ala86A (main-C) | hydrogen bond acceptor |
| Asp145A | hydrogen bond acceptor |
| His90A | hydrogen bond acceptor, hydrogen bond donor |
| Phe64A (main-N) | hydrogen bond donor |
| His90A | proton donor |
| Asp145A | electrofuge, proton acceptor |




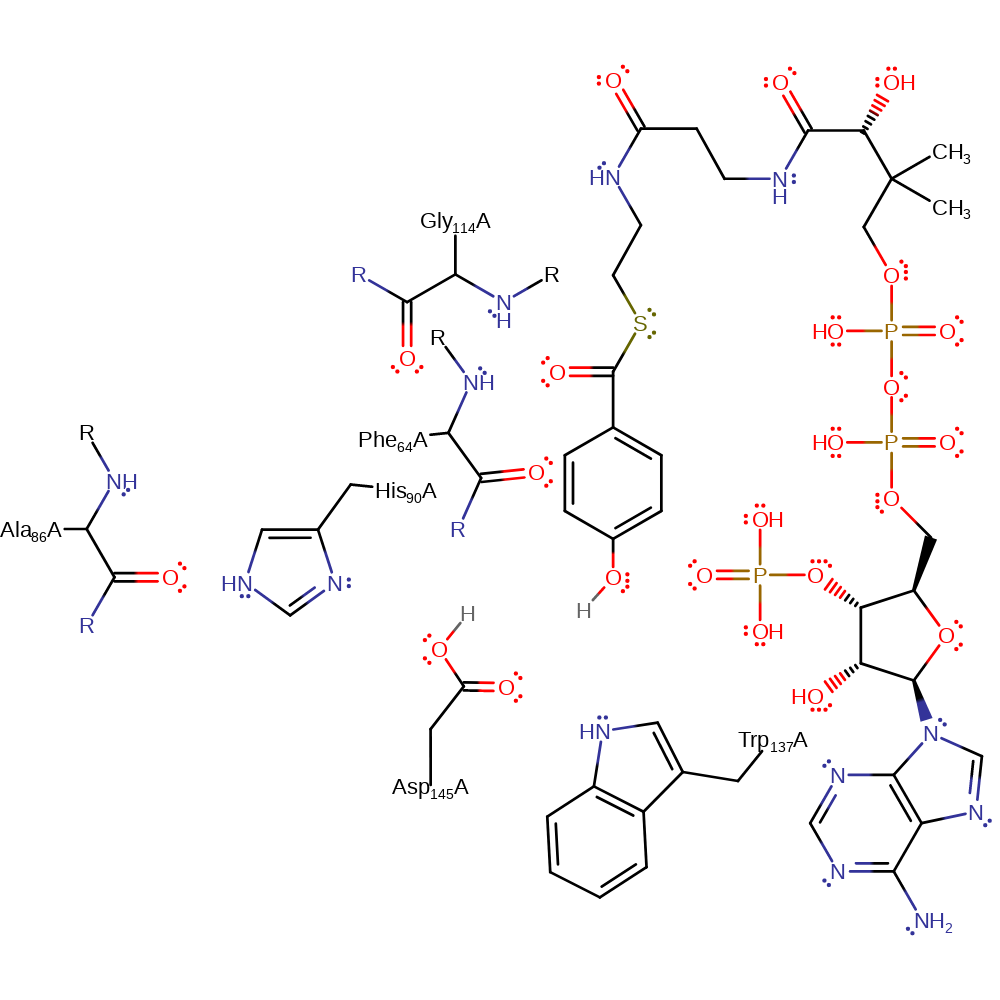 Download:
Download: