 |
PDBsum entry 5mdh

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
5mdh
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 1:
|
 |
E.C.1.1.1.37
- malate dehydrogenase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Citric acid cycle
|
 |
 |
 |
 |
 |
Reaction:
|
 |
(S)-malate + NAD+ = oxaloacetate + NADH + H+
|
 |
 |
 |
 |
 |
(S)-malate
Bound ligand (Het Group name = )
matches with 54.55% similarity
|
+
|
NAD(+)
Bound ligand (Het Group name = )
corresponds exactly
|
=
|
 oxaloacetate
oxaloacetate
|
+
|
 NADH
NADH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.1.1.1.96
- diiodophenylpyruvate reductase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
3-(3,5-diiodo-4-hydroxyphenyl)lactate + NAD+ = 3-(3,5-diiodo-4- hydroxyphenyl)pyruvate + NADH + H+
|
 |
 |
 |
 |
 |
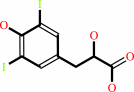 3-(3,5-diiodo-4-hydroxyphenyl)lactate
3-(3,5-diiodo-4-hydroxyphenyl)lactate
|
+
|
NAD(+)
Bound ligand (Het Group name = )
corresponds exactly
|
=
|
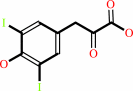 3-(3,5-diiodo-4- hydroxyphenyl)pyruvate
3-(3,5-diiodo-4- hydroxyphenyl)pyruvate
|
+
|
 NADH
NADH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Mol Biol
285:703-712
(1999)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structural basis of substrate specificity in malate dehydrogenases: crystal structure of a ternary complex of porcine cytoplasmic malate dehydrogenase, alpha-ketomalonate and tetrahydoNAD.
|
|
A.D.Chapman,
A.Cortés,
T.R.Dafforn,
A.R.Clarke,
R.L.Brady.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The structural basis for the extreme discrimination achieved by malate
dehydrogenases between a variety of closely related substrates encountered
within the cell has been difficult to assess because of the lack of an
appropriate catalytically competent structure of the enzyme. Here, we have
determined the crystal structure of a ternary complex of porcine cytoplasmic
malate dehydrogenase with the alternative substrate alpha-ketomalonate and the
coenzyme analogue 1,4,5,6-tetrahydronicotinamide. Both subunits of the dimeric
porcine heart, and from the prokaryotes Escherichia coli and Thermus flavus.
However, large changes are noted around the active site, where a mobile loop now
closes to bring key residues into contact with the substrate. This observation
substantiates a postulated mechanism in which the enzyme achieves high levels of
substrate discrimination through charge balancing in the active site. As the
activated cofactor/substrate complex has a net negative charge, a positive
counter-charge is provided by a conserved arginine in the active site loop. The
enzyme must, however, also discriminate against smaller substrates, such as
pyruvate. The structure shows in the closed (loop down) catalytically competent
complex two arginine residues (91 and 97) are driven into close proximity.
Without the complimentary, negative charge of the substrate side-chain of
oxaloacetate or alpha-ketomalonate, charge repulsion would resist formation
production of this catalytically productive conformation, hence minimising the
effectiveness of pyruvate as a substrate. By this mechanism, malate
dehydrogenase uses charge balancing to achieve fivefold orders of magnitude in
discrimination between potential substrates.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 7.
Figure 7. The position of the α-ketomalonate substrate
within the active site. The electron density was generated by
calculating 2|F[obs]|−|F[calc]| difference maps contoured at
1.3σ.
|
 |
Figure 8.
Figure 8. The hydrogen-bonding interactions of
α-ketomalonate with the key active site residues. This Figure
was generated using the program LIGPLOT [Wallace et al 1995].
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from Elsevier:
J Mol Biol
(1999,
285,
703-712)
copyright 1999.
|
|
| |
Figures were
selected
by the author.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
H.Xia,
C.Wu,
Q.Xu,
J.Shi,
F.Feng,
K.Chen,
Q.Yao,
Y.Wang,
and
L.Wang
(2011).
Molecular cloning and characterization of lactate dehydrogenase gene 1 in the silkworm, Bombyx mori.
|
| |
Mol Biol Rep,
38,
1853-1860.
|
 |
|
|
|
|
 |
Z.D.Wang,
B.J.Wang,
Y.D.Ge,
W.Pan,
J.Wang,
L.Xu,
A.M.Liu,
and
G.P.Zhu
(2011).
Expression and identification of a thermostable malate dehydrogenase from multicellular prokaryote Streptomyces avermitilis MA-4680.
|
| |
Mol Biol Rep,
38,
1629-1636.
|
 |
|
|
|
|
 |
J.D.Keighron,
and
C.D.Keating
(2010).
Enzyme:nanoparticle bioconjugates with two sequential enzymes: stoichiometry and activity of malate dehydrogenase and citrate synthase on Au nanoparticles.
|
| |
Langmuir,
26,
18992-19000.
|
 |
|
|
|
|
 |
A.Pradhan,
P.Mukherjee,
A.K.Tripathi,
M.A.Avery,
L.A.Walker,
and
B.L.Tekwani
(2009).
Analysis of quaternary structure of a [LDH-like] malate dehydrogenase of Plasmodium falciparum with oligomeric mutants.
|
| |
Mol Cell Biochem,
325,
141-148.
|
 |
|
|
|
|
 |
N.Zheng,
B.Huang,
J.Xu,
S.Huang,
J.Chen,
X.Hu,
K.Ying,
and
X.Yu
(2006).
Enzymatic and physico-chemical characteristics of recombinant cMDH and mMDH of Clonorchis sinensis.
|
| |
Parasitol Res,
99,
174-180.
|
 |
|
|
|
|
 |
B.Cox,
M.M.Chit,
T.Weaver,
C.Gietl,
J.Bailey,
E.Bell,
and
L.Banaszak
(2005).
Organelle and translocatable forms of glyoxysomal malate dehydrogenase. The effect of the N-terminal presequence.
|
| |
FEBS J,
272,
643-654.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
A.Cameron,
J.Read,
R.Tranter,
V.J.Winter,
R.B.Sessions,
R.L.Brady,
L.Vivas,
A.Easton,
H.Kendrick,
S.L.Croft,
D.Barros,
J.L.Lavandera,
J.J.Martin,
F.Risco,
S.García-Ochoa,
F.J.Gamo,
L.Sanz,
L.Leon,
J.R.Ruiz,
R.Gabarró,
A.Mallo,
and
F.Gómez de las Heras
(2004).
Identification and activity of a series of azole-based compounds with lactate dehydrogenase-directed anti-malarial activity.
|
| |
J Biol Chem,
279,
31429-31439.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
A.K.Tripathi,
P.V.Desai,
A.Pradhan,
S.I.Khan,
M.A.Avery,
L.A.Walker,
and
B.L.Tekwani
(2004).
An alpha-proteobacterial type malate dehydrogenase may complement LDH function in Plasmodium falciparum. Cloning and biochemical characterization of the enzyme.
|
| |
Eur J Biochem,
271,
3488-3502.
|
 |
|
|
|
|
 |
J.A.Lodge,
T.Maier,
W.Liebl,
V.Hoffmann,
and
N.Sträter
(2003).
Crystal structure of Thermotoga maritima alpha-glucosidase AglA defines a new clan of NAD+-dependent glycosidases.
|
| |
J Biol Chem,
278,
19151-19158.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
J.A.Read,
V.J.Winter,
C.M.Eszes,
R.B.Sessions,
and
R.L.Brady
(2001).
Structural basis for altered activity of M- and H-isozyme forms of human lactate dehydrogenase.
|
| |
Proteins,
43,
175-185.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
D.Madern
(2000).
The putative L-lactate dehydrogenase from Methanococcus jannaschii is an NADPH-dependent L-malate dehydrogenase.
|
| |
Mol Microbiol,
37,
1515-1520.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

