 |
PDBsum entry 1upf

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Transferase
|
 |
|
Title:
|
 |
Structure of the uracil phosphoribosyltransferase, mutant c128v bound to the drug 5-fluorouracil
|
|
Structure:
|
 |
Uracil phosphoribosyltransferase. Chain: d, c, b, a. Synonym: uprtase. Engineered: yes. Mutation: yes
|
|
Source:
|
 |
Toxoplasma gondii. Organism_taxid: 5811. Expressed in: escherichia coli. Expression_system_taxid: 562
|
|
Biol. unit:
|
 |
Homo-Dimer (from PDB file)
|
|
Resolution:
|
 |
|
|
Authors:
|
 |
M.A.Schumacher,D.Carter,D.Scott,D.Roos,B.Ullman,R.G.Brennan
|
Key ref:

|
 |
M.A.Schumacher
et al.
(1998).
Crystal structures of Toxoplasma gondii uracil phosphoribosyltransferase reveal the atomic basis of pyrimidine discrimination and prodrug binding.
EMBO J,
17,
3219-3232.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
17-Jun-98
|
Release date:
|
22-Jun-99
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
Q26998
(UPP_TOXGO) -
Uracil phosphoribosyltransferase from Toxoplasma gondii
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
244 a.a.
224 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|
*
PDB and UniProt seqs differ
at 3 residue positions (black
crosses)
|
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.4.2.9
- uracil phosphoribosyltransferase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
UMP + diphosphate = 5-phospho-alpha-D-ribose 1-diphosphate + uracil
|
 |
 |
 |
 |
 |
 UMP
UMP
|
+
|
 diphosphate
diphosphate
|
=
|
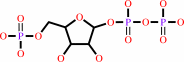 5-phospho-alpha-D-ribose 1-diphosphate
5-phospho-alpha-D-ribose 1-diphosphate
|
+
|
uracil
Bound ligand (Het Group name = )
matches with 88.89% similarity
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
EMBO J
17:3219-3232
(1998)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structures of Toxoplasma gondii uracil phosphoribosyltransferase reveal the atomic basis of pyrimidine discrimination and prodrug binding.
|
|
M.A.Schumacher,
D.Carter,
D.M.Scott,
D.S.Roos,
B.Ullman,
R.G.Brennan.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Uracil phosphoribosyltransferase (UPRTase) catalyzes the transfer of a ribosyl
phosphate group from alpha-D-5-phosphoribosyl-1-pyrophosphate to the N1 nitrogen
of uracil. The UPRTase from the opportunistic pathogen Toxoplasma gondii is a
rational target for antiparasitic drug design. To aid in structure-based drug
design studies against toxoplasmosis, the crystal structures of the T.gondii apo
UPRTase (1.93 A resolution), the UPRTase bound to its substrate, uracil (2.2 A
resolution), its product, UMP (2.5 A resolution), and the prodrug,
5-fluorouracil (2.3 A resolution), have been determined. These structures reveal
that UPRTase recognizes uracil through polypeptide backbone hydrogen bonds to
the uracil exocyclic O2 and endocyclic N3 atoms and a backbone-water-exocyclic
O4 oxygen hydrogen bond. This stereochemical arrangement and the architecture of
the uracil-binding pocket reveal why cytosine and pyrimidines with exocyclic
substituents at ring position 5 larger than fluorine, including thymine, cannot
bind to the enzyme. Strikingly, the T. gondii UPRTase contains a 22 residue
insertion within the conserved PRTase fold that forms an extended antiparallel
beta-arm. Leu92, at the tip of this arm, functions to cap the active site of its
dimer mate, thereby inhibiting the escape of the substrate-binding water
molecule.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 3.
Figure 3 Dimerization interface involving the  -arm.
Shown are the contacts made by the -arm.
Shown are the contacts made by the  -arm
(colored yellow) from one subunit to its dimer pair (colored
blue). Only part of the -arm
(colored yellow) from one subunit to its dimer pair (colored
blue). Only part of the  -arm
is shown for clarity. Labeled are those residues making dimer
contacts, and these include Phe83 and Phe101, which stack
against the N-terminus of A2', Tyr96 which is sandwiched between
Arg46' and Arg53', and Thr90 which makes hydrogen bonds to the
carbonyls of Pro231' and Gly232'. Also shown is Leu92, which
encloses the active site of the other monomer. Enzyme-bound
uracil and phosphate are displayed and colored by atom type,
whereby red is oxygen, blue is nitrogen, gray is carbon, and
magenta is phosphorous. -arm
is shown for clarity. Labeled are those residues making dimer
contacts, and these include Phe83 and Phe101, which stack
against the N-terminus of A2', Tyr96 which is sandwiched between
Arg46' and Arg53', and Thr90 which makes hydrogen bonds to the
carbonyls of Pro231' and Gly232'. Also shown is Leu92, which
encloses the active site of the other monomer. Enzyme-bound
uracil and phosphate are displayed and colored by atom type,
whereby red is oxygen, blue is nitrogen, gray is carbon, and
magenta is phosphorous.
|
 |
Figure 5.
Figure 5 Stereo view of the superimposition of UMP-bound UPRTase
(red) and 5-fluorouracil-bound UPRTase (green) onto the
uracil-bound UPRTase (blue). Note the rotation of the
5-fluorouracil ring that occurs to prevent steric clash between
the fluorine atom and the C[  ]atom
of Ala168. Also, the slightly 'pulled out' position of the
uracil ring of the UMP moiety, compared with uracil, is
revealed. Labeled are residues Ala168, Tyr228, which stacks over
the uracil ring, and Asp235 which hydrogen bonds with the
Tyr228. Wat1 of each complex is shown as an appropriately
colored sphere. The double-headed arrow highlights the proximity
of the fluorine atom of 5-fluorouracil to the C[ ]atom
of Ala168. Also, the slightly 'pulled out' position of the
uracil ring of the UMP moiety, compared with uracil, is
revealed. Labeled are residues Ala168, Tyr228, which stacks over
the uracil ring, and Asp235 which hydrogen bonds with the
Tyr228. Wat1 of each complex is shown as an appropriately
colored sphere. The double-headed arrow highlights the proximity
of the fluorine atom of 5-fluorouracil to the C[  ]atom
of Ala168. This figure was generated using O (Jones et al.,
1991). ]atom
of Ala168. This figure was generated using O (Jones et al.,
1991).
|
 |
|
|

|
| |
The above figures are
reprinted
from an Open Access publication published by Macmillan Publishers Ltd:
EMBO J
(1998,
17,
3219-3232)
copyright 1998.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
J.E.Hyde
(2007).
Targeting purine and pyrimidine metabolism in human apicomplexan parasites.
|
| |
Curr Drug Targets,
8,
31-47.
|
 |
|
|
|
|
 |
F.Arsène-Ploetze,
H.Nicoloff,
B.Kammerer,
J.Martinussen,
and
F.Bringel
(2006).
Uracil salvage pathway in Lactobacillus plantarum: Transcription and genetic studies.
|
| |
J Bacteriol,
188,
4777-4786.
|
 |
|
|
|
|
 |
K.A.Kantardjieff,
C.Vasquez,
P.Castro,
N.M.Warfel,
B.S.Rho,
T.Lekin,
C.Y.Kim,
B.W.Segelke,
T.C.Terwilliger,
and
B.Rupp
(2005).
Structure of pyrR (Rv1379) from Mycobacterium tuberculosis: a persistence gene and protein drug target.
|
| |
Acta Crystallogr D Biol Crystallogr,
61,
355-364.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
K.F.Jensen,
S.Arent,
S.Larsen,
and
L.Schack
(2005).
Allosteric properties of the GTP activated and CTP inhibited uracil phosphoribosyltransferase from the thermoacidophilic archaeon Sulfolobus solfataricus.
|
| |
FEBS J,
272,
1440-1453.
|
 |
|
|
|
|
 |
M.Kukimoto-Niino,
R.Shibata,
K.Murayama,
H.Hamana,
M.Nishimoto,
Y.Bessho,
T.Terada,
M.Shirouzu,
S.Kuramitsu,
and
S.Yokoyama
(2005).
Crystal structure of a predicted phosphoribosyltransferase (TT1426) from Thermus thermophilus HB8 at 2.01 A resolution.
|
| |
Protein Sci,
14,
823-827.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
A.R.Dodgson,
K.J.Dodgson,
C.Pujol,
M.A.Pfaller,
and
D.R.Soll
(2004).
Clade-specific flucytosine resistance is due to a single nucleotide change in the FUR1 gene of Candida albicans.
|
| |
Antimicrob Agents Chemother,
48,
2223-2227.
|
 |
|
|
|
|
 |
K.Chaudhary,
J.A.Darling,
L.M.Fohl,
W.J.Sullivan,
R.G.Donald,
E.R.Pfefferkorn,
B.Ullman,
and
D.S.Roos
(2004).
Purine salvage pathways in the apicomplexan parasite Toxoplasma gondii.
|
| |
J Biol Chem,
279,
31221-31227.
|
 |
|
|
|
|
 |
W.W.Hope,
L.Tabernero,
D.W.Denning,
and
M.J.Anderson
(2004).
Molecular mechanisms of primary resistance to flucytosine in Candida albicans.
|
| |
Antimicrob Agents Chemother,
48,
4377-4386.
|
 |
|
|
|
|
 |
A.Kadziola,
J.Neuhard,
and
S.Larsen
(2002).
Structure of product-bound Bacillus caldolyticus uracil phosphoribosyltransferase confirms ordered sequential substrate binding.
|
| |
Acta Crystallogr D Biol Crystallogr,
58,
936-945.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
C.Bashor,
J.M.Denu,
R.G.Brennan,
and
B.Ullman
(2002).
Kinetic mechanism of adenine phosphoribosyltransferase from Leishmania donovani.
|
| |
Biochemistry,
41,
4020-4031.
|
 |
|
|
|
|
 |
E.Kashuba,
V.Kashuba,
T.Sandalova,
G.Klein,
and
L.Szekely
(2002).
Epstein-Barr virus encoded nuclear protein EBNA-3 binds a novel human uridine kinase/uracil phosphoribosyltransferase.
|
| |
BMC Cell Biol,
3,
23.
|
 |
|
|
|
|
 |
H.Cao,
B.L.Pietrak,
and
C.Grubmeyer
(2002).
Quinolinate phosphoribosyltransferase: kinetic mechanism for a type II PRTase.
|
| |
Biochemistry,
41,
3520-3528.
|
 |
|
|
|
|
 |
M.A.Schumacher,
C.J.Bashor,
M.H.Song,
K.Otsu,
S.Zhu,
R.J.Parry,
B.Ullman,
and
R.G.Brennan
(2002).
The structural mechanism of GTP stabilized oligomerization and catalytic activation of the Toxoplasma gondii uracil phosphoribosyltransferase.
|
| |
Proc Natl Acad Sci U S A,
99,
78-83.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
D.C.McFadden,
M.Camps,
and
J.C.Boothroyd
(2001).
Resistance as a tool in the study of old and new drug targets in Toxoplasma.
|
| |
Drug Resist Updat,
4,
79-84.
|
 |
|
|
|
|
 |
E.R.Bonner,
J.N.D'Elia,
B.K.Billips,
and
R.L.Switzer
(2001).
Molecular recognition of pyr mRNA by the Bacillus subtilis attenuation regulatory protein PyrR.
|
| |
Nucleic Acids Res,
29,
4851-4865.
|
 |
|
|
|
|
 |
C.L.Phillips,
B.Ullman,
R.G.Brennan,
and
C.P.Hill
(1999).
Crystal structures of adenine phosphoribosyltransferase from Leishmania donovani.
|
| |
EMBO J,
18,
3533-3545.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
C.Lundegaard,
and
K.F.Jensen
(1999).
Kinetic mechanism of uracil phosphoribosyltransferase from Escherichia coli and catalytic importance of the conserved proline in the PRPP binding site.
|
| |
Biochemistry,
38,
3327-3334.
|
 |
|
|
|
|
 |
J.L.Smith
(1998).
Glutamine PRPP amidotransferase: snapshots of an enzyme in action.
|
| |
Curr Opin Struct Biol,
8,
686-694.
|
 |
|
|
|
|
 |
V.Sharma,
C.Grubmeyer,
and
J.C.Sacchettini
(1998).
Crystal structure of quinolinic acid phosphoribosyltransferase from Mmycobacterium tuberculosis: a potential TB drug target.
|
| |
Structure,
6,
1587-1599.
|
 |
|
PDB codes:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

