
|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Lyase (carbon-carbon)
|
 |
|
Title:
|
 |
Activated spinach rubisco complexed with 2-carboxyarabinitol bisphosphate
|
|
Structure:
|
 |
Ribulose-1,5-bisphosphate carboxylase/oxygenase. Chain: a, c, e, g. Synonym: rubisco. Ribulose-1,5-bisphosphate carboxylase/oxygenase. Chain: i, j, k, l. Synonym: rubisco. Ec: 4.1.1.39
|
|
Source:
|
 |
Spinacia oleracea. Spinach. Organism_taxid: 3562. Organ: leaf. Organ: leaf
|
|
Biol. unit:
|
 |
 60mer (from PDB file)
60mer (from PDB file)
|
|
Resolution:
|
 |
|
|
Authors:
|
 |
I.Andersson,S.Knight,C.-I.Branden
|
Key ref:

|
 |
I.Andersson
(1996).
Large structures at high resolution: the 1.6 A crystal structure of spinach ribulose-1,5-bisphosphate carboxylase/oxygenase complexed with 2-carboxyarabinitol bisphosphate.
J Mol Biol,
259,
160-174.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
22-Feb-96
|
Release date:
|
01-Aug-96
|
|
|
Supersedes:
|

|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
Chains A, C, E, G, I, J, K, L:
E.C.4.1.1.39
- ribulose-bisphosphate carboxylase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
2 (2R)-3-phosphoglycerate + 2 H+ = D-ribulose 1,5-bisphosphate + CO2 + H2O
|
 |
 |
 |
 |
 |
2
×
(2R)-3-phosphoglycerate
|
+
|
2
×
H(+)
|
=
|
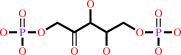 D-ribulose 1,5-bisphosphate
D-ribulose 1,5-bisphosphate
|
+
|
CO2
Bound ligand (Het Group name = )
matches with 85.71% similarity
|
+
|
H2O
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Mol Biol
259:160-174
(1996)
|
|
PubMed id:
|
|
|
|

|
| |
|
Large structures at high resolution: the 1.6 A crystal structure of spinach ribulose-1,5-bisphosphate carboxylase/oxygenase complexed with 2-carboxyarabinitol bisphosphate.
|
|
I.Andersson.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Ribulose-1,5-bisphosphate carboxylase/oxygenase (rubisco) from spinach is a
hexadecamer (L8S8, Mr = 550,000) consisting of eight large (L, 475 residues) and
eight small subunits (S, 123 residues). High-resolution data collection on
crystals with large unit cells is not a trivial task due to the effect of
radiation damage and the large number of overlapping reflections when
conventional data collection methods are used. In order to minimise these
effects, data on rubisco were collected with a giant Weissenberg camera at long
crystal to image-plate distances at the synchrotron of the Photon Factory,
Japan. Relative to conventional data sets, this experimental arrangement allowed
a 20 to 30-fold reduction of the X-ray dose/exposure time for data collection.
This paper describes the refined 1.6 A crystal structure of activated rubisco
complexed with a transition state analogue, 2-carboxyarabinitol-bisphosphate.
The crystallographic asymmetric unit contains an L4S4 unit, representing half of
the molecule. The structure presented here is currently the highest resolution
structure for any protein of comparable size. Refinement of the model was
carried out by restrained least squares techniques without non-crystallographic
symmetry averaging. The results show that all L and S subunits have identical
three-dimensional structures, and their arrangement within the hexadecamer has
no intrinsic asymmetry. A detailed analysis of the high-resolution maps
identified 30 differences in the sequence of the small subunit, indicating a
larger than usual heterogeneity for this nuclear encoded protein in spinach. No
such differences were found in the sequence of the chloroplast encoded large
subunit. The transition state analogue is in the cis conformation at the active
site suggesting a key role for the carbamate of Lys201 in catalysis. Analysis of
the active site around the catalytically essential magnesium ion further
indicates that residues in the second liganding sphere of the metal play a role
in fine-tuning the acid-base character and the position of the residues directly
liganded to the metal.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
Figure 1. Reactions catalysed by rubisco.
|
 |
Figure 7.
Figure 7. Overview of the active site of spinach rubisco showing 2-CABP, Mg
2+
and residues within hydrogen-bonding
distance to these ligands. The hydroxyl groups at C2 and C3 of 2-CABP are in cis conformation. The two views in (a)
and (b) are related by 180° with respect to the vertical axis.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from Elsevier:
J Mol Biol
(1996,
259,
160-174)
copyright 1996.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
H.Tamura,
Y.Saito,
H.Ashida,
Y.Kai,
T.Inoue,
A.Yokota,
and
H.Matsumura
(2009).
Structure of the apo decarbamylated form of 2,3-diketo-5-methylthiopentyl-1-phosphate enolase from Bacillus subtilis.
|
| |
Acta Crystallogr D Biol Crystallogr,
65,
942-951.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
S.Raunser,
R.Magnani,
Z.Huang,
R.L.Houtz,
R.C.Trievel,
P.A.Penczek,
and
T.Walz
(2009).
Rubisco in complex with Rubisco large subunit methyltransferase.
|
| |
Proc Natl Acad Sci U S A,
106,
3160-3165.
|
 |
|
|
|
|
 |
T.Genkov,
and
R.J.Spreitzer
(2009).
Highly conserved small subunit residues influence rubisco large subunit catalysis.
|
| |
J Biol Chem,
284,
30105-30112.
|
 |
|
|
|
|
 |
Y.Saito,
H.Ashida,
T.Sakiyama,
N.T.de Marsac,
A.Danchin,
A.Sekowska,
and
A.Yokota
(2009).
Structural and functional similarities between a ribulose-1,5-bisphosphate carboxylase/oxygenase (RuBisCO)-like protein from Bacillus subtilis and photosynthetic RuBisCO.
|
| |
J Biol Chem,
284,
13256-13264.
|
 |
|
|
|
|
 |
S.Satagopan,
and
R.J.Spreitzer
(2008).
Plant-like substitutions in the large-subunit carboxy terminus of Chlamydomonas Rubisco increase CO2/O2 Specificity.
|
| |
BMC Plant Biol,
8,
85.
|
 |
|
|
|
|
 |
F.R.Tabita,
T.E.Hanson,
H.Li,
S.Satagopan,
J.Singh,
and
S.Chan
(2007).
Function, structure, and evolution of the RubisCO-like proteins and their RubisCO homologs.
|
| |
Microbiol Mol Biol Rev,
71,
576-599.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
H.Li,
M.R.Sawaya,
F.R.Tabita,
and
D.Eisenberg
(2005).
Crystal structure of a RuBisCO-like protein from the green sulfur bacterium Chlorobium tepidum.
|
| |
Structure,
13,
779-789.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
R.J.Spreitzer,
S.R.Peddi,
and
S.Satagopan
(2005).
Phylogenetic engineering at an interface between large and small subunits imparts land-plant kinetic properties to algal Rubisco.
|
| |
Proc Natl Acad Sci U S A,
102,
17225-17230.
|
 |
|
|
|
|
 |
S.Satagopan,
and
R.J.Spreitzer
(2004).
Substitutions at the Asp-473 latch residue of chlamydomonas ribulosebisphosphate carboxylase/oxygenase cause decreases in carboxylation efficiency and CO(2)/O(2) specificity.
|
| |
J Biol Chem,
279,
14240-14244.
|
 |
|
|
|
|
 |
Y.C.Du,
S.R.Peddi,
and
R.J.Spreitzer
(2003).
Assessment of structural and functional divergence far from the large subunit active site of ribulose-1,5-bisphosphate carboxylase/oxygenase.
|
| |
J Biol Chem,
278,
49401-49405.
|
 |
|
|
|
|
 |
Y.Marcus,
H.Altman-Gueta,
A.Finkler,
and
M.Gurevitz
(2003).
Dual role of cysteine 172 in redox regulation of ribulose 1,5-bisphosphate carboxylase/oxygenase activity and degradation.
|
| |
J Bacteriol,
185,
1509-1517.
|
 |
|
|
|
|
 |
J.B.Utåker,
K.Andersen,
A.Aakra,
B.Moen,
and
I.F.Nes
(2002).
Phylogeny and functional expression of ribulose 1,5-bisphosphate carboxylase/oxygenase from the autotrophic ammonia-oxidizing bacterium Nitrosospira sp. isolate 40KI.
|
| |
J Bacteriol,
184,
468-478.
|
 |
|
|
|
|
 |
N.Maeda,
T.Kanai,
H.Atomi,
and
T.Imanaka
(2002).
The unique pentagonal structure of an archaeal Rubisco is essential for its high thermostability.
|
| |
J Biol Chem,
277,
31656-31662.
|
 |
|
|
|
|
 |
R.J.Spreitzer,
and
M.E.Salvucci
(2002).
Rubisco: structure, regulatory interactions, and possibilities for a better enzyme.
|
| |
Annu Rev Plant Biol,
53,
449-475.
|
 |
|
|
|
|
 |
K.Kitano,
N.Maeda,
T.Fukui,
H.Atomi,
T.Imanaka,
and
K.Miki
(2001).
Crystal structure of a novel-type archaeal rubisco with pentagonal symmetry.
|
| |
Structure,
9,
473-481.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
T.C.Taylor,
A.Backlund,
K.Bjorhall,
R.J.Spreitzer,
and
I.Andersson
(2001).
First crystal structure of Rubisco from a green alga, Chlamydomonas reinhardtii.
|
| |
J Biol Chem,
276,
48159-48164.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
Y.C.Du,
S.Hong,
and
R.J.Spreitzer
(2000).
RbcS suppressor mutations improve the thermal stability and CO2/O2 specificity of rbcL- mutant ribulose-1,5-bisphosphate carboxylase/oxygenase.
|
| |
Proc Natl Acad Sci U S A,
97,
14206-14211.
|
 |
|
|
|
|
 |
A.Mattevi,
G.Tedeschi,
L.Bacchella,
A.Coda,
A.Negri,
and
S.Ronchi
(1999).
Structure of L-aspartate oxidase: implications for the succinate dehydrogenase/fumarate reductase oxidoreductase family.
|
| |
Structure,
7,
745-756.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
E.L.Hegg,
A.K.Whiting,
R.E.Saari,
J.McCracken,
R.P.Hausinger,
and
L.Que
(1999).
Herbicide-degrading alpha-keto acid-dependent enzyme TfdA: metal coordination environment and mechanistic insights.
|
| |
Biochemistry,
38,
16714-16726.
|
 |
|
|
|
|
 |
H.Sugawara,
H.Yamamoto,
N.Shibata,
T.Inoue,
S.Okada,
C.Miyake,
A.Yokota,
and
Y.Kai
(1999).
Crystal structure of carboxylase reaction-oriented ribulose 1, 5-bisphosphate carboxylase/oxygenase from a thermophilic red alga, Galdieria partita.
|
| |
J Biol Chem,
274,
15655-15661.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
J.Moreno,
and
R.J.Spreitzer
(1999).
C172S substitution in the chloroplast-encoded large subunit affects stability and stress-induced turnover of ribulose-1,5-bisphosphate carboxylase/oxygenase.
|
| |
J Biol Chem,
274,
26789-26793.
|
 |
|
|
|
|
 |
R.Douce,
and
M.Neuburger
(1999).
Biochemical dissection of photorespiration.
|
| |
Curr Opin Plant Biol,
2,
214-222.
|
 |
|
|
|
|
 |
S.Khan,
P.J.Andralojc,
P.J.Lea,
and
M.A.Parry
(1999).
2'-carboxy-D-arabitinol 1-phosphate protects ribulose 1, 5-bisphosphate carboxylase/oxygenase against proteolytic breakdown.
|
| |
Eur J Biochem,
266,
840-847.
|
 |
|
|
|
|
 |
M.Hippler,
K.Redding,
and
J.D.Rochaix
(1998).
Chlamydomonas genetics, a tool for the study of bioenergetic pathways.
|
| |
Biochim Biophys Acta,
1367,
1.
|
 |
|
|
|
|
 |
M.R.Harpel,
F.W.Larimer,
and
F.C.Hartman
(1998).
Multiple catalytic roles of His 287 of Rhodospirillum rubrum ribulose 1,5-bisphosphate carboxylase/oxygenase.
|
| |
Protein Sci,
7,
730-738.
|
 |
|
|
|
|
 |
W.A.King,
J.E.Gready,
and
T.J.Andrews
(1998).
Quantum chemical analysis of the enolization of ribulose bisphosphate: the first hurdle in the fixation of CO2 by Rubisco.
|
| |
Biochemistry,
37,
15414-15422.
|
 |
|
|
|
|
 |
S.Hong,
and
R.J.Spreitzer
(1997).
Complementing substitutions at the bottom of the barrel influence catalysis and stability of ribulose-bisphosphate carboxylase/oxygenase.
|
| |
J Biol Chem,
272,
11114-11117.
|
 |
|
|
|
|
 |
T.C.Taylor,
and
I.Andersson
(1997).
Structure of a product complex of spinach ribulose-1,5-bisphosphate carboxylase/oxygenase.
|
| |
Biochemistry,
36,
4041-4046.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.R.Harpel,
and
F.C.Hartman
(1996).
Facilitation of the terminal proton transfer reaction of ribulose 1,5-bisphosphate carboxylase/oxygenase by active-site Lys166.
|
| |
Biochemistry,
35,
13865-13870.
|
 |
|
|
|
|
 |
N.Shibata,
T.Inoue,
K.Fukuhara,
Y.Nagara,
R.Kitagawa,
S.Harada,
N.Kasai,
K.Uemura,
K.Kato,
A.Yokota,
and
Y.Kai
(1996).
Orderly disposition of heterogeneous small subunits in D-ribulose-1,5-bisphosphate carboxylase/oxygenase from spinach.
|
| |
J Biol Chem,
271,
26449-26452.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
T.C.Taylor,
M.D.Fothergill,
and
I.Andersson
(1996).
A common structural basis for the inhibition of ribulose 1,5-bisphosphate carboxylase by 4-carboxyarabinitol 1,5-bisphosphate and xylulose 1,5-bisphosphate.
|
| |
J Biol Chem,
271,
32894-32899.
|
 |
|
PDB codes:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

