
|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Oxidoreductase
|
 |
|
Title:
|
 |
Activated spinach rubisco in complex with the product 3- phosphoglycerate
|
|
Structure:
|
 |
Ribulose bisphosphate carboxylase (large chain). Chain: l, b, e, h. Synonym: rubisco. Ribulose bisphosphate carboxylase (small chain). Chain: s, c, f, i. Synonym: rubisco. Ec: 4.1.1.39
|
|
Source:
|
 |
Spinacia oleracea. Spinach. Organism_taxid: 3562. Organ: leaf. Organ: leaf
|
|
Biol. unit:
|
 |
Octamer (from PDB file)
|
|
Resolution:
|
 |
|
2.20Å
|
R-factor:
|
0.207
|
R-free:
|
0.224
|
|
|
Authors:
|
 |
T.C.Taylor,I.Andersson
|
Key ref:

|
 |
T.C.Taylor
and
I.Andersson
(1997).
Structure of a product complex of spinach ribulose-1,5-bisphosphate carboxylase/oxygenase.
Biochemistry,
36,
4041-4046.
PubMed id:
DOI:
|
 |
|
Date:
|
 |
|
20-Jan-97
|
Release date:
|
07-Jul-97
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
Chains L, S, B, C, E, F, H, I:
E.C.4.1.1.39
- ribulose-bisphosphate carboxylase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
2 (2R)-3-phosphoglycerate + 2 H+ = D-ribulose 1,5-bisphosphate + CO2 + H2O
|
 |
 |
 |
 |
 |
2
×
(2R)-3-phosphoglycerate
|
+
|
2
×
H(+)
|
=
|
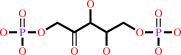 D-ribulose 1,5-bisphosphate
D-ribulose 1,5-bisphosphate
|
+
|
CO2
Bound ligand (Het Group name = )
matches with 52.63% similarity
|
+
|
H2O
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Biochemistry
36:4041-4046
(1997)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structure of a product complex of spinach ribulose-1,5-bisphosphate carboxylase/oxygenase.
|
|
T.C.Taylor,
I.Andersson.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The crystal structure of an activated complex of ribulose-1,5-bisphosphate
carboxylase/oxygenase from spinach and its product 3-phosphoglycerate has been
determined to 2.2 A resolution. The structure is of the open form with the
active site accessible to the solvent as observed in the structures of the
activated ligand-free enzyme and the complex of the activated enzyme with the
substrate ribulose-1,5-bisphosphate. Two molecules of 3-phosphoglycerate are
bound per active site. The phosphates of both molecules bind approximately at
the same position as the phosphates of ribulose-1,5-bisphosphate or the
six-carbon intermediate analogue 2-carboxyarabinitol-1,5-bisphosphate, but one
product molecule is swung out from the active site with its carboxylate group
pointing toward solution. The present structure points to direct participation
of the active site side chain of lysine 175 in later stages of catalysis. This
possibility is discussed in the light of mutagenesis studies.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
H.Tamura,
Y.Saito,
H.Ashida,
Y.Kai,
T.Inoue,
A.Yokota,
and
H.Matsumura
(2009).
Structure of the apo decarbamylated form of 2,3-diketo-5-methylthiopentyl-1-phosphate enolase from Bacillus subtilis.
|
| |
Acta Crystallogr D Biol Crystallogr,
65,
942-951.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
B.Pierce,
W.Tong,
and
Z.Weng
(2005).
M-ZDOCK: a grid-based approach for Cn symmetric multimer docking.
|
| |
Bioinformatics,
21,
1472-1478.
|
 |
|
|
|
|
 |
H.Li,
M.R.Sawaya,
F.R.Tabita,
and
D.Eisenberg
(2005).
Crystal structure of a RuBisCO-like protein from the green sulfur bacterium Chlorobium tepidum.
|
| |
Structure,
13,
779-789.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
R.J.Spreitzer,
and
M.E.Salvucci
(2002).
Rubisco: structure, regulatory interactions, and possibilities for a better enzyme.
|
| |
Annu Rev Plant Biol,
53,
449-475.
|
 |
|
|
|
|
 |
K.Kitano,
N.Maeda,
T.Fukui,
H.Atomi,
T.Imanaka,
and
K.Miki
(2001).
Crystal structure of a novel-type archaeal rubisco with pentagonal symmetry.
|
| |
Structure,
9,
473-481.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
K.Motohashi,
A.Kondoh,
M.T.Stumpp,
and
T.Hisabori
(2001).
Comprehensive survey of proteins targeted by chloroplast thioredoxin.
|
| |
Proc Natl Acad Sci U S A,
98,
11224-11229.
|
 |
|
|
|
|
 |
T.C.Taylor,
A.Backlund,
K.Bjorhall,
R.J.Spreitzer,
and
I.Andersson
(2001).
First crystal structure of Rubisco from a green alga, Chlamydomonas reinhardtii.
|
| |
J Biol Chem,
276,
48159-48164.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
C.Dabney-Smith,
P.W.van Den Wijngaard,
Y.Treece,
W.J.Vredenberg,
and
B.D.Bruce
(1999).
The C terminus of a chloroplast precursor modulates its interaction with the translocation apparatus and PIRAC.
|
| |
J Biol Chem,
274,
32351-32359.
|
 |
|
|
|
|
 |
H.Ishida,
A.Makino,
and
T.Mae
(1999).
Fragmentation of the large subunit of ribulose-1,5-bisphosphate carboxylase by reactive oxygen species occurs near Gly-329.
|
| |
J Biol Chem,
274,
5222-5226.
|
 |
|
|
|
|
 |
H.Sugawara,
H.Yamamoto,
N.Shibata,
T.Inoue,
S.Okada,
C.Miyake,
A.Yokota,
and
Y.Kai
(1999).
Crystal structure of carboxylase reaction-oriented ribulose 1, 5-bisphosphate carboxylase/oxygenase from a thermophilic red alga, Galdieria partita.
|
| |
J Biol Chem,
274,
15655-15661.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
M.R.Harpel,
F.W.Larimer,
and
F.C.Hartman
(1998).
Multiple catalytic roles of His 287 of Rhodospirillum rubrum ribulose 1,5-bisphosphate carboxylase/oxygenase.
|
| |
Protein Sci,
7,
730-738.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

