 |
PDBsum entry 1geh
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Lyase
|
 |
|
Title:
|
 |
Crystal structure of archaeal rubisco (ribulose 1,5-bisphosphate carboxylase/oxygenase)
|
|
Structure:
|
 |
Ribulose-1,5-bisphosphate carboxylase/oxygenase. Chain: a, b, c, d, e. Engineered: yes
|
|
Source:
|
 |
Thermococcus kodakarensis. Organism_taxid: 69014. Strain: kod1. Expressed in: escherichia coli. Expression_system_taxid: 562
|
|
Biol. unit:
|
 |
 Decamer (from PDB file)
Decamer (from PDB file)
|
|
Resolution:
|
 |
|
2.80Å
|
R-factor:
|
0.224
|
R-free:
|
0.254
|
|
|
Authors:
|
 |
K.Kitano,N.Maeda,T.Fukui,H.Atomi,T.Imanaka,K.Miki
|
Key ref:

|
 |
K.Kitano
et al.
(2001).
Crystal structure of a novel-type archaeal rubisco with pentagonal symmetry.
Structure,
9,
473-481.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
13-Nov-00
|
Release date:
|
19-Dec-01
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
O93627
(RBL_THEKO) -
Ribulose bisphosphate carboxylase from Thermococcus kodakarensis (strain ATCC BAA-918 / JCM 12380 / KOD1)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
444 a.a.
427 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.4.1.1.39
- ribulose-bisphosphate carboxylase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
2 (2R)-3-phosphoglycerate + 2 H+ = D-ribulose 1,5-bisphosphate + CO2 + H2O
|
 |
 |
 |
 |
 |
2
×
(2R)-3-phosphoglycerate
|
+
|
2
×
H(+)
|
=
|
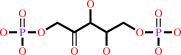 D-ribulose 1,5-bisphosphate
D-ribulose 1,5-bisphosphate
|
+
|
 CO2
CO2
|
+
|
H2O
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Structure
9:473-481
(2001)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structure of a novel-type archaeal rubisco with pentagonal symmetry.
|
|
K.Kitano,
N.Maeda,
T.Fukui,
H.Atomi,
T.Imanaka,
K.Miki.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
BACKGROUND: Ribulose 1,5-bisphosphate carboxylase/oxygenase (Rubisco) is the key
enzyme of the Calvin-Benson cycle and catalyzes the primary reaction of CO2
fixation in plants, algae, and bacteria. Rubiscos have been so far classified
into two types. Type I is composed of eight large subunits (L subunits) and
eight small subunits (S subunits) with tetragonal symmetry (L8S8), but type II
is usually composed only of two L subunits (L2). Recently, some genuinely active
Rubiscos of unknown physiological function have been reported from archaea.
RESULTS: The crystal structure of Rubisco from the hyperthermophilic archaeon
Thermococcus kodakaraensis KOD1 (Tk-Rubisco) was determined at 2.8 A resolution.
The enzyme is composed only of L subunits and showed a novel (L2)5 decameric
structure. Compared to previously known type I enzymes, each L2 dimer is
inclined approximately 16 degrees to form a toroid-shaped decamer with its
unique L2-L2 interfaces. Differential scanning calorimetry (DSC), circular
dichroism (CD), and gel permeation chromatography (GPC) showed that Tk-Rubisco
maintains its secondary structure and decameric assembly even at high
temperatures. CONCLUSIONS: The present study provides the first structure of an
archaeal Rubisco, an unprecedented (L2)5 decamer. Biochemical studies indicate
that Tk-Rubisco maintains its decameric structure at high temperatures. The
structure is distinct from type I and type II Rubiscos and strongly supports
that Tk-Rubisco should be classified as a novel type III Rubisco.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |
Figure 4.
Figure 4. Oligomeric Structures of Rubiscos(a) Decamer
structure of Tk-Rubisco, with ribbon diagram from top view. Each
monomer is shown in different colors and labeled from "A" to
"J." Plausible structures of RuBP molecules and magnesium ions
are marked in ellipses.(b) Side view of Tk-Rubisco by -90°
rotation from (a), with the AB dimer at the center.(c) The
(L[2])[4] core of type I Rubisco, with ribbon diagram from top
view, where only L subunits in the L[8]S[8] structure [11] are
shown. Each monomer is shown in different colors. RuBP molecules
and magnesium ions are marked in ellipses.(d) Side view of the
(L[2])[4] core of type I Rubisco by -90° rotation from (c)

|
 |
|
|

|
| |
The above figure is
reprinted
by permission from Cell Press:
Structure
(2001,
9,
473-481)
copyright 2001.
|
|
| |
Figure was
selected
by the author.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
H.Alonso,
M.J.Blayney,
J.L.Beck,
and
S.M.Whitney
(2009).
Substrate-induced assembly of Methanococcoides burtonii D-ribulose-1,5-bisphosphate carboxylase/oxygenase dimers into decamers.
|
| |
J Biol Chem,
284,
33876-33882.
|
 |
|
|
|
|
 |
S.Satagopan,
S.S.Scott,
T.G.Smith,
and
F.R.Tabita
(2009).
A Rubisco mutant that confers growth under a normally "inhibitory" oxygen concentration.
|
| |
Biochemistry,
48,
9076-9083.
|
 |
|
|
|
|
 |
F.R.Tabita,
T.E.Hanson,
S.Satagopan,
B.H.Witte,
and
N.E.Kreel
(2008).
Phylogenetic and evolutionary relationships of RubisCO and the RubisCO-like proteins and the functional lessons provided by diverse molecular forms.
|
| |
Philos Trans R Soc Lond B Biol Sci,
363,
2629-2640.
|
 |
|
|
|
|
 |
A.R.Portis,
and
M.A.Parry
(2007).
Discoveries in Rubisco (Ribulose 1,5-bisphosphate carboxylase/oxygenase): a historical perspective.
|
| |
Photosynth Res,
94,
121-143.
|
 |
|
|
|
|
 |
H.E.Elsaied,
H.Kimura,
and
T.Naganuma
(2007).
Composition of archaeal, bacterial, and eukaryal RuBisCO genotypes in three Western Pacific arc hydrothermal vent systems.
|
| |
Extremophiles,
11,
191-202.
|
 |
|
|
|
|
 |
O.Mueller-Cajar,
and
M.R.Badger
(2007).
New roads lead to Rubisco in archaebacteria.
|
| |
Bioessays,
29,
722-724.
|
 |
|
|
|
|
 |
S.Yoshida,
H.Atomi,
and
T.Imanaka
(2007).
Engineering of a type III rubisco from a hyperthermophilic archaeon in order to enhance catalytic performance in mesophilic host cells.
|
| |
Appl Environ Microbiol,
73,
6254-6261.
|
 |
|
|
|
|
 |
T.Sato,
H.Atomi,
and
T.Imanaka
(2007).
Archaeal type III RuBisCOs function in a pathway for AMP metabolism.
|
| |
Science,
315,
1003-1006.
|
 |
|
|
|
|
 |
A.Carré-Mlouka,
A.Méjean,
P.Quillardet,
H.Ashida,
Y.Saito,
A.Yokota,
I.Callebaut,
A.Sekowska,
E.Dittmann,
C.Bouchier,
and
N.T.de Marsac
(2006).
A new rubisco-like protein coexists with a photosynthetic rubisco in the planktonic cyanobacteria Microcystis.
|
| |
J Biol Chem,
281,
24462-24471.
|
 |
|
|
|
|
 |
H.Li,
M.R.Sawaya,
F.R.Tabita,
and
D.Eisenberg
(2005).
Crystal structure of a RuBisCO-like protein from the green sulfur bacterium Chlorobium tepidum.
|
| |
Structure,
13,
779-789.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
M.W.Finn,
and
F.R.Tabita
(2003).
Synthesis of catalytically active form III ribulose 1,5-bisphosphate carboxylase/oxygenase in archaea.
|
| |
J Bacteriol,
185,
3049-3059.
|
 |
|
|
|
|
 |
H.Atomi
(2002).
Microbial enzymes involved in carbon dioxide fixation.
|
| |
J Biosci Bioeng,
94,
497-505.
|
 |
|
|
|
|
 |
H.Imanaka,
T.Fukui,
H.Atomi,
and
T.Imanaka
(2002).
Gene cloning and characterization of fructose-1,6-bisphosphate aldolase from the hyperthermophilic archaeon Thermococcus kodakaraensis KOD1.
|
| |
J Biosci Bioeng,
94,
237-243.
|
 |
|
|
|
|
 |
N.Maeda,
T.Kanai,
H.Atomi,
and
T.Imanaka
(2002).
The unique pentagonal structure of an archaeal Rubisco is essential for its high thermostability.
|
| |
J Biol Chem,
277,
31656-31662.
|
 |
|
|
|
|
 |
T.Fukui,
T.Eguchi,
H.Atomi,
and
T.Imanaka
(2002).
A membrane-bound archaeal Lon protease displays ATP-independent proteolytic activity towards unfolded proteins and ATP-dependent activity for folded proteins.
|
| |
J Bacteriol,
184,
3689-3698.
|
 |
|
|
|
|
 |
T.Imanaka,
and
H.Atomi
(2002).
Catalyzing "hot" reactions: enzymes from hyperthermophilic Archaea.
|
| |
Chem Rec,
2,
149-163.
|
 |
|
|
|
|
 |
T.C.Taylor,
A.Backlund,
K.Bjorhall,
R.J.Spreitzer,
and
I.Andersson
(2001).
First crystal structure of Rubisco from a green alga, Chlamydomonas reinhardtii.
|
| |
J Biol Chem,
276,
48159-48164.
|
 |
|
PDB code:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

