 |
PDBsum entry 5onh
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Ligase
|
 |
|
Title:
|
 |
Quaternary complex of wild type e. Coli leucyl-tRNA synthetase with tRNA(leu), leucyl-adenylate analogue, and post-transfer editing analogue of norvaline in the aminoacylation conformation
|
|
Structure:
|
 |
Leucine--tRNA ligase. Chain: a, d. Synonym: leucyl-tRNA synthetase,leurs. Engineered: yes. L-leucyl-tRNA(leu). Chain: b, e. Engineered: yes
|
|
Source:
|
 |
Escherichia coli k-12. Organism_taxid: 83333. Gene: leus, b0642, jw0637. Expressed in: escherichia coli bl21. Expression_system_taxid: 511693. Synthetic: yes. Escherichia coli. Organism_taxid: 562
|
|
Resolution:
|
 |
|
3.10Å
|
R-factor:
|
0.224
|
R-free:
|
0.257
|
|
|
Authors:
|
 |
A.Palencia,S.Cusack
|
|
Key ref:
|
 |
M.Dulic
et al.
(2018).
Kinetic Origin of Substrate Specificity in Post-Transfer Editing by Leucyl-tRNA Synthetase.
J Mol Biol,
430,
1.
PubMed id:
DOI:
|
 |
|
Date:
|
 |
|
03-Aug-17
|
Release date:
|
15-Nov-17
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P07813
(SYL_ECOLI) -
Leucine--tRNA ligase from Escherichia coli (strain K12)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
860 a.a.
860 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
|

|
|
|

|

|
|
|
|
G-C-C-C-G-G-A-U-G-G-U-G-G-A-A-U-C-G-G-U-A-G-A-C-A-C-A-A-G-G-G-A-U-U-A-A-U-C-C-
82 bases
|
|
|
|
G-C-C-C-G-G-A-U-G-G-U-G-G-A-A-U-C-G-G-U-A-G-A-C-A-C-A-A-G-G-G-A-U-A-A-U-C-C-C-
83 bases
|
|

|
 |
 |
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.6.1.1.4
- leucine--tRNA ligase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
tRNA(Leu) + L-leucine + ATP = L-leucyl-tRNA(Leu) + AMP + diphosphate
|
 |
 |
 |
 |
 |
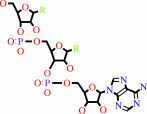 tRNA(Leu)
tRNA(Leu)
|
+
|
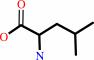 L-leucine
L-leucine
|
+
|
 ATP
ATP
|
=
|
L-leucyl-tRNA(Leu)
Bound ligand (Het Group name = )
matches with 54.29% similarity
|
+
|
 AMP
AMP
|
+
|
 diphosphate
diphosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Mol Biol
430:1
(2018)
|
|
PubMed id:
|
|
|
|

|
| |
|
Kinetic Origin of Substrate Specificity in Post-Transfer Editing by Leucyl-tRNA Synthetase.
|
|
M.Dulic,
N.Cvetesic,
I.Zivkovic,
A.Palencia,
S.Cusack,
B.Bertosa,
I.Gruic-Sovulj.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The intrinsic editing capacities of aminoacyl-tRNA synthetases ensure a
high-fidelity translation of the amino acids that possess effective non-cognate
aminoacylation surrogates. The dominant error-correction pathway comprises
deacylation of misaminoacylated tRNA within the aminoacyl-tRNA synthetase
editing site. To assess the origin of specificity of Escherichia coli
leucyl-tRNA synthetase (LeuRS) against the cognate aminoacylation product in
editing, we followed binding and catalysis independently using cognate leucyl-
and non-cognate norvalyl-tRNALeuand their non-hydrolyzable analogues.
We found that the amino acid part (leucine versus norvaline) of
(mis)aminoacyl-tRNAs can contribute approximately 10-fold to ground-state
discrimination at the editing site. In sharp contrast, the rate of deacylation
of leucyl- and norvalyl-tRNALeudiffered by about 104-fold.
We further established the critical role for the A76 3'-OH group of the
tRNALeuin post-transfer editing, which supports the
substrate-assisted deacylation mechanism. Interestingly, the abrogation of the
LeuRS specificity determinant threonine 252 did not improve the affinity of the
editing site for the cognate leucine as expected, but instead substantially
enhanced the rate of leucyl-tRNALeuhydrolysis. In line with that,
molecular dynamics simulations revealed that the wild-type enzyme, but not the
T252A mutant, enforced leucine to adopt the side-chain conformation that
promotes the steric exclusion of a putative catalytic water. Our data
demonstrated that the LeuRS editing site exhibits amino acid specificity of
kinetic origin, arguing against the anticipated prominent role of steric
exclusion in the rejection of leucine. This feature distinguishes editing from
the synthetic site, which relies on ground-state discrimination in amino acid
selection.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

