 |
PDBsum entry 5odc

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
5odc
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|

|

|

|

|

|

 653 a.a.
653 a.a.
|
 |

|

|

|

|

|

|

|

 291 a.a.
291 a.a.
|
 |

|

|

|

|

|

|

|
 184 a.a.
184 a.a.
|
 |

|

|

|

|

|

|

|

 138 a.a.
138 a.a.
|
 |

|

|

|

|

|

|

|

 298 a.a.
298 a.a.
|
 |

|

|

|

|

|

|

|

 447 a.a.
447 a.a.
|
 |

|

|

|

|

|

|

|
 173 a.a.
173 a.a.
|
 |

|

|
|
|
|
|
|
|
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Oxidoreductase
|
 |
|
Title:
|
 |
Heterodisulfide reductase / [nife]-hydrogenase complex from methanothermococcus thermolithotrophicus at 2.3 a resolution
|
|
Structure:
|
 |
Heterodisulfide reductase, subunit a. Chain: a, g. Other_details: gb ref wp_018154264.1. Heterodisulfide reductase, subunit b. Chain: b, h. Other_details: gb ref wp_018154154.1, this subunit contains two pentacoordinated non cubane [4fe-4s] cluster. Heterodisulfide reductase, subunit c. Chain: c, i.
|
|
Source:
|
 |
Methanothermococcus thermolithotrophicus dsm 2095. Organism_taxid: 523845. Cell_line: /. Atcc: /. Organ: /. Tissue: /. Tissue: /
|
|
Resolution:
|
 |
|
2.30Å
|
R-factor:
|
0.179
|
R-free:
|
0.200
|
|
|
Authors:
|
 |
T.Wagner,J.Koch,U.Ermler,S.Shima
|
|
Key ref:
|
 |
T.Wagner
et al.
(2017).
Methanogenic heterodisulfide reductase (HdrABC-MvhAGD) uses two noncubane [4Fe-4S] clusters for reduction.
Science,
357,
699-703.
PubMed id:
|
 |
|
Date:
|
 |
|
05-Jul-17
|
Release date:
|
30-Aug-17
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
A0A2D0TCB9
(A0A2D0TCB9_METTL) -
CoB--CoM heterodisulfide reductase iron-sulfur subunit A from Methanothermococcus thermolithotrophicus DSM 2095
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
654 a.a.
653 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|

|

|
|
A0A2D0TCB4
(A0A2D0TCB4_METTL) -
Heterodisulfide reductase, subunit B from Methanothermococcus thermolithotrophicus DSM 2095
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
291 a.a.
291 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|

|

|
|
A0A2D0TC97
(A0A2D0TC97_METTL) -
Heterodisulfide reductase, subunit C from Methanothermococcus thermolithotrophicus DSM 2095
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
184 a.a.
184 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|

|

|
|
A0A2D0TC98
(A0A2D0TC98_METTL) -
Methyl-viologen reducing hydrogenase subunit D from Methanothermococcus thermolithotrophicus DSM 2095
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
140 a.a.
138 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|

|

|
|
A0A2D0TC99
(A0A2D0TC99_METTL) -
Methyl-viologen reducing hydrogenase subunit G from Methanothermococcus thermolithotrophicus DSM 2095
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
299 a.a.
298 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
 |
|
|
 |
 |
 |
 |
Enzyme class 2:
|
 |
Chains A, G:
E.C.1.8.-.-
|
|
 |
 |
 |
 |
 |
Enzyme class 3:
|
 |
Chains B, C, D, H, I, J:
E.C.1.8.98.1
- dihydromethanophenazine:CoB--CoM heterodisulfide reductase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
|
 |
 |
 |
 |
 |
Reaction:
|
 |
methanophenazine + coenzyme B + coenzyme M = dihydromethanophenazine + coenzyme M-coenzyme B heterodisulfide
|
 |
 |
 |
 |
 |
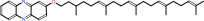 methanophenazine
methanophenazine
|
+
|
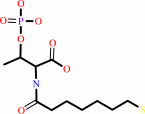 coenzyme B
coenzyme B
|
+
|
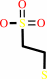 coenzyme M
coenzyme M
|
=
|
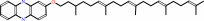 dihydromethanophenazine
dihydromethanophenazine
|
+
|
coenzyme M-coenzyme B heterodisulfide
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Heme; Iron-sulfur
|
 |
 |
 |
 |
 |
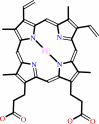 Heme
Heme
|
 Iron-sulfur
Iron-sulfur
|
|
 |
 |
Enzyme class 4:
|
 |
Chains E, F, K, L:
E.C.1.12.1.2
- hydrogen dehydrogenase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
H2 + NAD+ = NADH + H+
|
 |
 |
 |
 |
 |
H2
|
+
|
 NAD(+)
NAD(+)
|
=
|
 NADH
NADH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
FAD or FMN; Iron-sulfur; Ni(2+)
|
 |
 |
 |
 |
 |
 FAD
FAD
|
|
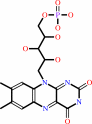 FMN
FMN
|
 Iron-sulfur
Iron-sulfur
|
Ni(2+)
|
|
 |
 |
Enzyme class 5:
|
 |
Chains E, F, K, L:
E.C.1.12.99.6
- hydrogenase (acceptor).
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
H2 + A = AH2
|
 |
 |
 |
 |
 |
Cofactor:
|
 |
Iron-sulfur; Ni(2+)
|
 |
 |
 |
 |
 |
 Iron-sulfur
Iron-sulfur
|
|
|
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
Science
357:699-703
(2017)
|
|
PubMed id:
|
|
|
|

|
| |
|
Methanogenic heterodisulfide reductase (HdrABC-MvhAGD) uses two noncubane [4Fe-4S] clusters for reduction.
|
|
T.Wagner,
J.Koch,
U.Ermler,
S.Shima.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
In methanogenic archaea, the carbon dioxide (CO2) fixation and
methane-forming steps are linked through the heterodisulfide reductase
(HdrABC)-[NiFe]-hydrogenase (MvhAGD) complex that uses flavin-based electron
bifurcation to reduce ferredoxin and the heterodisulfide of coenzymes M and B.
Here, we present the structure of the native heterododecameric HdrABC-MvhAGD
complex at 2.15-angstrom resolution. HdrB contains two noncubane [4Fe-4S]
clusters composed of fused [3Fe-4S]-[2Fe-2S] units sharing 1 iron (Fe) and 1
sulfur (S), which were coordinated at the CCG motifs. Soaking experiments showed
that the heterodisulfide is clamped between the two noncubane [4Fe-4S] clusters
and homolytically cleaved, forming coenzyme M and B bound to each iron.
Coenzymes are consecutively released upon one-by-one electron transfer. The
HdrABC-MvhAGD atomic model serves as a structural template for numerous HdrABC
homologs involved in diverse microbial metabolic pathways.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |

| | |

