 |
PDBsum entry 5ja2
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 2:
|
 |
Chain A:
E.C.6.2.1.72
- L-serine--[L-seryl-carrier protein] ligase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
holo-[peptidyl-carrier protein] + L-serine + ATP = L-seryl-[peptidyl- carrier protein] + AMP + diphosphate
|
 |
 |
 |
 |
 |
holo-[peptidyl-carrier protein]
|
+
|
 L-serine
L-serine
|
+
|
ATP
Bound ligand (Het Group name = )
matches with 44.44% similarity
|
=
|
L-seryl-[peptidyl- carrier protein]
|
+
|
 AMP
AMP
|
+
|
 diphosphate
diphosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 3:
|
 |
Chain A:
E.C.6.3.2.14
- enterobactin synthase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
3 2,3-dihydroxybenzoate + 3 L-serine + 6 ATP = enterobactin + 6 AMP + 6 diphosphate + 4 H+
|
 |
 |
 |
 |
 |
3
×
2,3-dihydroxybenzoate
Bound ligand (Het Group name = )
matches with 54.55% similarity
|
+
|
 3
×
L-serine
3
×
L-serine
|
+
|
 6
×
ATP
6
×
ATP
|
=
|
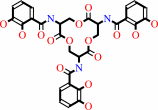 enterobactin
enterobactin
|
+
|
 6
×
AMP
6
×
AMP
|
+
|
 6
×
diphosphate
6
×
diphosphate
|
+
|
4
×
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Biol Chem
291:22559-22571
(2016)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structures of a Nonribosomal Peptide Synthetase Module Bound to MbtH-like Proteins Support a Highly Dynamic Domain Architecture.
|
|
B.R.Miller,
E.J.Drake,
C.Shi,
C.C.Aldrich,
A.M.Gulick.
|
|
|

|
| |
ABSTRACT
|
|

|
| |
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|

