 |
PDBsum entry 5e9f
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Lyase
|
 |
|
Title:
|
 |
Structural insights of isocitrate lyases from magnaporthe oryzae
|
|
Structure:
|
 |
Isocitrate lyase. Chain: a, b, c, d. Synonym: icl,isocitrase,isocitratase,methylisocitrate lyase,mica, threo-d(s)-isocitrate glyoxylate-lyase. Engineered: yes
|
|
Source:
|
 |
Magnaporthe oryzae 70-15. Rice blast fungus. Organism_taxid: 242507. Strain: 70-15. Gene: icl1, mgg_04895. Expressed in: escherichia coli. Expression_system_taxid: 562.
|
|
Resolution:
|
 |
|
2.80Å
|
R-factor:
|
0.187
|
R-free:
|
0.226
|
|
|
Authors:
|
 |
Y.Park,Y.Cho,Y.-H.Lee,Y.-W.Lee,S.Rhee
|
|
Key ref:
|
 |
Y.Park
et al.
(2016).
Crystal structure and functional analysis of isocitrate lyases from Magnaporthe oryzae and Fusarium graminearum.
J Struct Biol,
194,
395-403.
PubMed id:
DOI:
|
 |
|
Date:
|
 |
|
15-Oct-15
|
Release date:
|
27-Apr-16
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 2:
|
 |
Chains A, B, C, D:
E.C.4.1.3.1
- isocitrate lyase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Glyoxylate Cycle
|
 |
 |
 |
 |
 |
Reaction:
|
 |
D-threo-isocitrate = glyoxylate + succinate
|
 |
 |
 |
 |
 |
D-threo-isocitrate
|
=
|
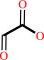 glyoxylate
glyoxylate
|
+
|
 succinate
succinate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 3:
|
 |
Chains A, B, C, D:
E.C.4.1.3.30
- methylisocitrate lyase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
(2S,3R)-3-hydroxybutane-1,2,3-tricarboxylate = pyruvate + succinate
|
 |
 |
 |
 |
 |
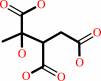 (2S,3R)-3-hydroxybutane-1,2,3-tricarboxylate
(2S,3R)-3-hydroxybutane-1,2,3-tricarboxylate
|
=
|
 pyruvate
pyruvate
|
+
|
 succinate
succinate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Struct Biol
194:395-403
(2016)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structure and functional analysis of isocitrate lyases from Magnaporthe oryzae and Fusarium graminearum.
|
|
Y.Park,
Y.Cho,
Y.H.Lee,
Y.W.Lee,
S.Rhee.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The glyoxylate cycle bypasses a CO2-generating step in the tricarboxylic acid
(TCA) cycle and efficiently assimilates C2 compounds into intermediates that can
be used in later steps of the TCA cycle. It plays an essential role in pathogen
survival during host infection such that the enzymes involved in this cycle have
been suggested as potential drug targets against human pathogens. Isocitrate
lyase (ICL) catalyzes the first-step reaction of the glyoxylate cycle, using
isocitrate from the TCA cycle as the substrate to produce succinate and
glyoxylate. In this study we report the crystal structure of Magnaporthe oryzae
ICL in both the ligand-free form and as a complex with Mg(2+), glyoxylate, and
glycerol, as well as the structure of the Fusarium graminearum ICL complexed
with Mn(2+) and malonate. We also describe the ligand-induced conformational
changes in the catalytic loop and C-terminal region, both of which are essential
for catalysis. Using various mutant ICLs in an activity assay, we gained insight
into the function of residues within the active site. These structural and
functional analyses provide detailed information with regard to fungal ICLs.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|

