 |
PDBsum entry 5af7
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.3.13.1.4
- 3-sulfinopropanoyl-CoA desulfinase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
3-sulfinopropanoyl-CoA + H2O = propanoyl-CoA + sulfite + H+
|
 |
 |
 |
 |
 |
3-sulfinopropanoyl-CoA
|
+
|
H2O
|
=
|
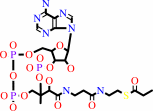 propanoyl-CoA
propanoyl-CoA
|
+
|
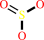 sulfite
sulfite
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
FAD
|
 |
 |
 |
 |
 |
FAD
Bound ligand (Het Group name =
FAD)
corresponds exactly
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Acta Crystallogr D Biol Crystallogr
71:1360-1372
(2015)
|
|
PubMed id:
|
|
|
|

|
| |
|
3-Sulfinopropionyl-coenzyme A (3SP-CoA) desulfinase from Advenella mimigardefordensis DPN7(T): crystal structure and function of a desulfinase with an acyl-CoA dehydrogenase fold.
|
|
M.Schürmann,
R.Meijers,
T.R.Schneider,
A.Steinbüchel,
M.Cianci.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
3-Sulfinopropionyl-coenzyme A (3SP-CoA) desulfinase (AcdDPN7; EC 3.13.1.4) was
identified during investigation of the 3,3'-dithiodipropionic acid (DTDP)
catabolic pathway in the betaproteobacterium Advenella mimigardefordensis strain
DPN7(T). DTDP is an organic disulfide and a precursor for the synthesis of
polythioesters (PTEs) in bacteria, and is of interest for biotechnological PTE
production. AcdDPN7 catalyzes sulfur abstraction from 3SP-CoA, a key step during
the catabolism of DTDP. Here, the crystal structures of apo AcdDPN7 at 1.89 Å
resolution and of its complex with the CoA moiety from the substrate analogue
succinyl-CoA at 2.30 Å resolution are presented. The apo structure shows that
AcdDPN7 belongs to the acyl-CoA dehydrogenase superfamily fold and that it is a
tetramer, with each subunit containing one flavin adenine dinucleotide (FAD)
molecule. The enzyme does not show any dehydrogenase activity. Dehydrogenase
activity would require a catalytic base (Glu or Asp residue) at either position
246 or position 366, where a glutamine and a glycine are instead found,
respectively, in this desulfinase. The positioning of CoA in the crystal complex
enabled the modelling of a substrate complex containing 3SP-CoA. This indicates
that Arg84 is a key residue in the desulfination reaction. An Arg84Lys mutant
showed a complete loss of enzymatic activity, suggesting that the guanidinium
group of the arginine is essential for desulfination. AcdDPN7 is the first
desulfinase with an acyl-CoA dehydrogenase fold to be reported, which underlines
the versatility of this enzyme scaffold.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

