 |
PDBsum entry 3vms
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Transferase
|
 |
|
Title:
|
 |
Crystal structure of staphylococcus aureus membrane-bound transglycosylase in complex with nbd-lipid ii
|
|
Structure:
|
 |
Monofunctional glycosyltransferase. Chain: a, b. Fragment: residues 28-269. Synonym: mgt, peptidoglycan tgase. Engineered: yes
|
|
Source:
|
 |
Staphylococcus aureus. Organism_taxid: 158878. Strain: mu50. Gene: mgt. Expressed in: escherichia coli. Expression_system_taxid: 562.
|
|
Resolution:
|
 |
|
3.20Å
|
R-factor:
|
0.265
|
R-free:
|
0.330
|
|
|
Authors:
|
 |
C.Y.Huang,H.W.Shih,L.Y.Lin,Y.W.Tien,T.J.R.Cheng,W.C.Cheng,C.H.Wong, C.Ma
|
|
Key ref:
|
 |
C.Y.Huang
et al.
(2012).
Crystal structure of Staphylococcus aureus transglycosylase in complex with a lipid II analog and elucidation of peptidoglycan synthesis mechanism.
Proc Natl Acad Sci U S A,
109,
6496-6501.
PubMed id:
|
 |
|
Date:
|
 |
|
15-Dec-11
|
Release date:
|
18-Apr-12
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
Q99T05
(MGT_STAAM) -
Monofunctional glycosyltransferase from Staphylococcus aureus (strain Mu50 / ATCC 700699)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
269 a.a.
220 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.4.1.129
- Transferred entry: 2.4.99.28.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Peptidoglycan Biosynthesis (Part 3)
|
 |
 |
 |
 |
 |
Reaction:
|
 |
[GlcNAc-(1->4)-Mur2Ac(oyl-L-Ala-gamma-D-Glu-L-Lys-D-Ala-D-Ala)](n)- di-trans,octa-cis-undecaprenyl diphosphate + beta-D-GlcNAc-(1->4)- Mur2Ac(oyl-L-Ala-gamma-D-Glu-L-Lys-D-Ala-D-Ala)-di-trans,octa- cis-undecaprenyl diphosphate = [GlcNAc-(1->4)-Mur2Ac(oyl-L-Ala-gamma-D- Glu-L-Lys-D-Ala-D-Ala)](n+1)-di-trans-octa-cis-undecaprenyl diphosphate + di-trans,octa-cis-undecaprenyl diphosphate + H+
|
 |
 |
 |
 |
 |
[GlcNAc-(1->4)-Mur2Ac(oyl-L-Ala-gamma-D-Glu-L-Lys-D-Ala-D-Ala)](n)- di-trans,octa-cis-undecaprenyl diphosphate
|
+
|
beta-D-GlcNAc-(1->4)- Mur2Ac(oyl-L-Ala-gamma-D-Glu-L-Lys-D-Ala-D-Ala)-di-trans,octa- cis-undecaprenyl diphosphate
|
=
|
[GlcNAc-(1->4)-Mur2Ac(oyl-L-Ala-gamma-D- Glu-L-Lys-D-Ala-D-Ala)](n+1)-di-trans-octa-cis-undecaprenyl diphosphate
|
+
|
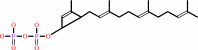 di-trans,octa-cis-undecaprenyl diphosphate
di-trans,octa-cis-undecaprenyl diphosphate
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
Proc Natl Acad Sci U S A
109:6496-6501
(2012)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structure of Staphylococcus aureus transglycosylase in complex with a lipid II analog and elucidation of peptidoglycan synthesis mechanism.
|
|
C.Y.Huang,
H.W.Shih,
L.Y.Lin,
Y.W.Tien,
T.J.Cheng,
W.C.Cheng,
C.H.Wong,
C.Ma.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Bacterial transpeptidase and transglycosylase on the surface are essential for
cell wall synthesis, and many antibiotics have been developed to target the
transpeptidase; however, the problem of antibiotic resistance has arisen and
caused a major threat in bacterial infection. The transglycosylase has been
considered to be another excellent target, but no antibiotics have been
developed to target this enzyme. Here, we determined the crystal structure of
the Staphylococcus aureus membrane-bound transglycosylase, monofunctional
glycosyltransferase, in complex with a lipid II analog to 2.3 Å resolution. Our
results showed that the lipid II-contacting residues are not only conserved in
WT and drug-resistant bacteria but also significant in enzymatic activity.
Mechanistically, we proposed that K140 and R148 in the donor site, instead of
the previously proposed E156, are used to stabilize the pyrophosphate-leaving
group of lipid II, and E100 in the acceptor site acts as general base for the
4-OH of GlcNAc to facilitate the transglycosylation reaction. This mechanism,
further supported by mutagenesis study and the structure of monofunctional
glycosyltransferase in complex with moenomycin in the donor site, provides a
direction for antibacterial drugs design.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

