 |
PDBsum entry 3spd

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Hydrolase/DNA
|
PDB id
|
|

|

|
3spd
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Hydrolase/DNA
|
 |
|
Title:
|
 |
Crystal structure of aprataxin ortholog hnt3 in complex with DNA
|
|
Structure:
|
 |
Aprataxin-like protein. Chain: a, b, c, d. Fragment: unp residues 33-232. Synonym: hnt3 protein, hit family protein 3. Engineered: yes. Mutation: yes. DNA (5'-d( Tp Ap Tp Tp Cp Cp Gp Ap Tp Ap Gp Tp Gp Ap C)- 3'). Chain: e, g.
|
|
Source:
|
 |
Schizosaccharomyces pombe. Fission yeast. Organism_taxid: 284812. Strain: atcc 38366 / 972. Gene: hnt3, spcc18.09c. Expressed in: escherichia coli. Expression_system_taxid: 562. Synthetic: yes. Other_details: DNA oligonucleotides were synthesized from shanghai
|
|
Resolution:
|
 |
|
1.91Å
|
R-factor:
|
0.177
|
R-free:
|
0.212
|
|
|
Authors:
|
 |
Y.Gong,D.Zhu,J.Ding,C.Dou,X.Ren,T.Jiang,D.Wang
|
|
Key ref:
|
 |
Y.Gong
et al.
(2011).
Crystal structures of aprataxin ortholog Hnt3 reveal the mechanism for reversal of 5'-adenylated DNA.
Nat Struct Biol,
18,
1297-1299.
PubMed id:
|
 |
|
Date:
|
 |
|
01-Jul-11
|
Release date:
|
12-Oct-11
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
O74859
(APTX_SCHPO) -
Aprataxin-like protein from Schizosaccharomyces pombe (strain 972 / ATCC 24843)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
232 a.a.
200 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|
*
PDB and UniProt seqs differ
at 1 residue position (black
cross)
|
|

|
|
|

|

|
|
|
|
T-A-T-T-C-C-G-A-T-A-G-T-G-A
14 bases
|
|
|
|
G-T-C-A-C-T-A-T-C-G-G-A-A-T
14 bases
|
|
|
|
T-A-T-T-C-C-G-A-T-A-G-T-G-A
14 bases
|
|
|
|
G-T-C-A-C-T-A-T-C-G-G-A-A-T
14 bases
|
|

|
 |
 |
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.3.6.1.71
- adenosine-5'-diphospho-5'-[DNA] diphosphatase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
|
1.
|
a 5'-end adenosine-5'-diphospho-5'-2'-deoxyribonucleoside-DNA + H2O = a 5'-end 5'-phospho-2'-deoxyribonucleoside-DNA + AMP + 2 H+
|
|
2.
|
a 5'-end adenosine-5'-diphospho-5'-ribonucleoside- 2'-deoxyribonucleotide-DNA + H2O = a 5'-end 5'-phospho-ribonucleoside- 2'-deoxyribonucleotide-DNA + AMP + 2 H+
|
|
 |
 |
 |
 |
 |
5'-end adenosine-5'-diphospho-5'-2'-deoxyribonucleoside-DNA
|
+
|
H2O
|
=
|
5'-end 5'-phospho-2'-deoxyribonucleoside-DNA
|
+
|
 AMP
AMP
|
+
|
2
×
H(+)
|
|
 |
 |
 |
 |
 |
5'-end adenosine-5'-diphospho-5'-ribonucleoside- 2'-deoxyribonucleotide-DNA
|
+
|
H2O
|
=
|
5'-end 5'-phospho-ribonucleoside- 2'-deoxyribonucleotide-DNA
|
+
|
 AMP
AMP
|
+
|
2
×
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 3:
|
 |
E.C.3.6.1.72
- DNA-3'-diphospho-5'-guanosine diphosphatase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
a 3'-end 2'-deoxyribonucleotide-3'-diphospho-5'-guanosine-DNA + H2O = a 3'-end 2'-deoxyribonucleotide 3'-phosphate-DNA + GMP + 2 H+
|
 |
 |
 |
 |
 |
3'-end 2'-deoxyribonucleotide-3'-diphospho-5'-guanosine-DNA
|
+
|
H2O
|
=
|
3'-end 2'-deoxyribonucleotide 3'-phosphate-DNA
|
+
|
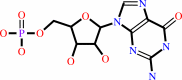 GMP
GMP
|
+
|
2
×
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
Nat Struct Biol
18:1297-1299
(2011)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structures of aprataxin ortholog Hnt3 reveal the mechanism for reversal of 5'-adenylated DNA.
|
|
Y.Gong,
D.Zhu,
J.Ding,
C.N.Dou,
X.Ren,
L.Gu,
T.Jiang,
D.C.Wang.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Aprataxin is a DNA deadenylase that resolves DNA 5'-AMP termini and reverses
abortive DNA ligation. The crystal structures of Schizosaccharomyces pombe
aprataxin Hnt3 in its apo form and in complex to dsDNA and dsDNA-AMP reveal how
Hnt3 recognizes and processes 5'-adenylated DNA in a structure-specific manner.
The bound DNA adopts a 5'-flap conformation that facilitates 5'-AMP access to
the active site, where AMP cleavage occurs by a canonical catalytic mechanism.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

