 |
PDBsum entry 3nqd

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Lyase/lyase inhibitor
|
PDB id
|
|

|

|
3nqd
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Lyase/lyase inhibitor
|
 |
|
Title:
|
 |
Crystal structure of the mutant i96t of orotidine 5'-monophosphate decarboxylase from methanobacterium thermoautotrophicum complexed with inhibitor bmp
|
|
Structure:
|
 |
Orotidine 5'-phosphate decarboxylase. Chain: a, b. Synonym: omp decarboxylase, ompdcase, ompdecase. Engineered: yes. Mutation: yes
|
|
Source:
|
 |
Methanothermobacter thermautotrophicus. Organism_taxid: 145262. Gene: pyrf, mth_129. Expressed in: escherichia coli. Expression_system_taxid: 562
|
|
Resolution:
|
 |
|
1.42Å
|
R-factor:
|
0.166
|
R-free:
|
0.185
|
|
|
Authors:
|
 |
A.A.Fedorov,E.V.Fedorov,B.M.Wood,J.A.Gerlt,S.C.Almo
|
|
Key ref:
|
 |
V.Iiams
et al.
(2011).
Mechanism of the orotidine 5'-monophosphate decarboxylase-catalyzed reaction: importance of residues in the orotate binding site.
Biochemistry,
50,
8497-8507.
PubMed id:
|
 |
|
Date:
|
 |
|
29-Jun-10
|
Release date:
|
11-May-11
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
O26232
(PYRF_METTH) -
Orotidine 5'-phosphate decarboxylase from Methanothermobacter thermautotrophicus (strain ATCC 29096 / DSM 1053 / JCM 10044 / NBRC 100330 / Delta H)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
228 a.a.
219 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|
*
PDB and UniProt seqs differ
at 2 residue positions (black
crosses)
|
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.4.1.1.23
- orotidine-5'-phosphate decarboxylase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Pyrimidine Biosynthesis
|
 |
 |
 |
 |
 |
Reaction:
|
 |
orotidine 5'-phosphate + H+ = UMP + CO2
|
 |
 |
 |
 |
 |
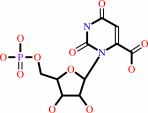 orotidine 5'-phosphate
orotidine 5'-phosphate
|
+
|
H(+)
|
=
|
 UMP
UMP
|
+
|
CO2
Bound ligand (Het Group name = )
matches with 95.45% similarity
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
Biochemistry
50:8497-8507
(2011)
|
|
PubMed id:
|
|
|
|

|
| |
|
Mechanism of the orotidine 5'-monophosphate decarboxylase-catalyzed reaction: importance of residues in the orotate binding site.
|
|
V.Iiams,
B.J.Desai,
A.A.Fedorov,
E.V.Fedorov,
S.C.Almo,
J.A.Gerlt.
|
|
|

|
| |
ABSTRACT
|
|

|
| |
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

