 |
PDBsum entry 3hcc

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.1.1.28
- phenylethanolamine N-methyltransferase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Dopa Biosynthesis
|
 |
 |
 |
 |
 |
Reaction:
|
 |
phenylethanolamine + S-adenosyl-L-methionine = N-methylphenylethanolamine + S-adenosyl-L-homocysteine + H+
|
 |
 |
 |
 |
 |
phenylethanolamine
Bound ligand (Het Group name = )
matches with 52.94% similarity
|
+
|
 S-adenosyl-L-methionine
S-adenosyl-L-methionine
|
=
|
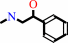 N-methylphenylethanolamine
N-methylphenylethanolamine
|
+
|
 S-adenosyl-L-homocysteine
S-adenosyl-L-homocysteine
|
+
|
H(+)
Bound ligand (Het Group name = )
corresponds exactly
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
Biochem J
422:463-471
(2009)
|
|
PubMed id:
|
|
|
|

|
| |
|
Molecular recognition of physiological substrate noradrenaline by the adrenaline-synthesizing enzyme PNMT and factors influencing its methyltransferase activity.
|
|
N.Drinkwater,
C.L.Gee,
M.Puri,
K.R.Criscione,
M.J.McLeish,
G.L.Grunewald,
J.L.Martin.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Substrate specificity is critically important for enzyme catalysis. In the
adrenaline-synthesizing enzyme PNMT (phenylethanolamine N-methyltransferase),
minor changes in substituents can convert substrates into inhibitors. Here we
report the crystal structures of six human PNMT complexes, including the first
structure of the enzyme in complex with its physiological ligand
R-noradrenaline. Determining this structure required rapid soak methods because
of the tendency for noradrenaline to oxidize. Comparison of the
PNMT-noradrenaline complex with the previously determined PNMT-p-octopamine
complex demonstrates that these two substrates form almost equivalent
interactions with the enzyme and show that p-octopamine is a valid model
substrate for PNMT. The crystal structures illustrate the adaptability of the
PNMT substrate binding site in accepting multi-fused ring systems, such as
substituted norbornene, as well as noradrenochrome, the oxidation product of
noradrenaline. These results explain why only a subset of ligands recognized by
PNMT are methylated by the enzyme; bulky substituents dictate the binding
orientation of the ligand and can thereby place the acceptor amine too far from
the donor methyl group for methylation to occur. We also show how the critical
Glu(185) catalytic residue can be replaced by aspartic acid with a loss of only
10-fold in catalytic efficiency. This is because protein backbone movements
place the Asp(185) carboxylate almost coincident with the carboxylate of
Glu(185). Conversely, replacement of Glu(185) by glutamine reduces catalytic
efficiency almost 300-fold, not only because of the loss of charge, but also
because the variant residue does not adopt the same conformation as Glu(185).

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
A.K.Malde,
and
A.E.Mark
(2011).
Challenges in the determination of the binding modes of non-standard ligands in X-ray crystal complexes.
|
| |
J Comput Aided Mol Des,
25,
1.
|
 |
|
|
|
|
 |
N.Drinkwater,
H.Vu,
K.M.Lovell,
K.R.Criscione,
B.M.Collins,
T.E.Prisinzano,
S.A.Poulsen,
M.J.McLeish,
G.L.Grunewald,
and
J.L.Martin
(2010).
Fragment-based screening by X-ray crystallography, MS and isothermal titration calorimetry to identify PNMT (phenylethanolamine N-methyltransferase) inhibitors.
|
| |
Biochem J,
431,
51-61.
|
 |
|
PDB codes:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

