 |
PDBsum entry 3kpv

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.1.1.28
- phenylethanolamine N-methyltransferase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Dopa Biosynthesis
|
 |
 |
 |
 |
 |
Reaction:
|
 |
phenylethanolamine + S-adenosyl-L-methionine = N-methylphenylethanolamine + S-adenosyl-L-homocysteine + H+
|
 |
 |
 |
 |
 |
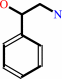 phenylethanolamine
phenylethanolamine
|
+
|
 S-adenosyl-L-methionine
S-adenosyl-L-methionine
|
=
|
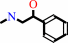 N-methylphenylethanolamine
N-methylphenylethanolamine
|
+
|
 S-adenosyl-L-homocysteine
S-adenosyl-L-homocysteine
|
+
|
H(+)
Bound ligand (Het Group name = )
corresponds exactly
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
Biochem J
431:51-61
(2010)
|
|
PubMed id:
|
|
|
|

|
| |
|
Fragment-based screening by X-ray crystallography, MS and isothermal titration calorimetry to identify PNMT (phenylethanolamine N-methyltransferase) inhibitors.
|
|
N.Drinkwater,
H.Vu,
K.M.Lovell,
K.R.Criscione,
B.M.Collins,
T.E.Prisinzano,
S.A.Poulsen,
M.J.McLeish,
G.L.Grunewald,
J.L.Martin.
|
|
|

|
| |
ABSTRACT
|
|

|
| |
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

