 |
PDBsum entry 3ctf

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
3ctf
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.1.11.1.9
- glutathione peroxidase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
2 glutathione + H2O2 = glutathione disulfide + 2 H2O
|
 |
 |
 |
 |
 |
 2
×
glutathione
2
×
glutathione
|
+
|
 H2O2
H2O2
|
=
|
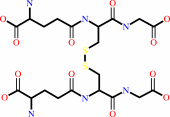 glutathione disulfide
glutathione disulfide
|
+
|
2
×
H2O
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Se(2+)
|
 |
 |
 |
 |
 |
Enzyme class 3:
|
 |
E.C.2.5.1.18
- glutathione transferase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
RX + glutathione = an S-substituted glutathione + a halide anion + H+
|
 |
 |
 |
 |
 |
 2
×
RX
2
×
RX
|
+
|
 glutathione
glutathione
|
=
|
S-substituted glutathione
|
+
|
2
×
halide anion
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
Biochim Biophys Acta
1804:1542-1547
(2010)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structural basis for the different activities of yeast Grx1 and Grx2.
|
|
W.F.Li,
J.Yu,
X.X.Ma,
Y.B.Teng,
M.Luo,
Y.J.Tang,
C.Z.Zhou.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Yeast glutaredoxins Grx1 and Grx2 catalyze the reduction of both inter- and
intra-molecular disulfide bonds using glutathione (GSH) as the electron donor.
Although sharing the same dithiolic CPYC active site and a sequence identity of
64%, they have been proved to play different roles during oxidative stress and
to possess different glutathione-disulfide reductase activities. To address the
structural basis of these differences, we solved the crystal structures of Grx2
in oxidized and reduced forms, at 2.10 A and 1.50 A, respectively. With the Grx1
structures we previously reported, comparative structural analyses revealed that
Grx1 and Grx2 share a similar GSH binding site, except for a single residue
substitution from Asp89 in Grx1 to Ser123 in Grx2. Site-directed mutagenesis in
combination with activity assays further proved this single residue variation is
critical for the different activities of yeast Grx1 and Grx2.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

