
|

|

|
|
 |
Contents |
 |
|
|

|

|

|

|

|

|

|
 485 a.a.
485 a.a.
|
 |

|

|

|

|

|

|

|
 398 a.a.
398 a.a.
|
 |

|

|

|

|

|

|

|
 99 a.a.
99 a.a.
|
 |

|

|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Ligase
|
 |
|
Title:
|
 |
Structure of tRNA-dependent amidotransferase gatcab
|
|
Structure:
|
 |
Glutamyl-tRNA(gln) amidotransferase subunit a. Chain: a. Synonym: glu-adt subunit a. Engineered: yes. Aspartyl/glutamyl-tRNA(asn/gln) amidotransferase subunit b. Chain: b. Synonym: asp/glu-adt subunit b. Engineered: yes. Aspartyl/glutamyl-tRNA(asn/gln) amidotransferase subunit c.
|
|
Source:
|
 |
Staphylococcus aureus. Organism_taxid: 1280. Strain: mu50. Gene: gata. Expressed in: escherichia coli. Expression_system_taxid: 562. Gene: gatb. Gene: gatc.
|
|
Biol. unit:
|
 |
Trimer (from
)
|
|
Resolution:
|
 |
|
2.50Å
|
R-factor:
|
0.238
|
R-free:
|
0.275
|
|
|
Authors:
|
 |
A.Nakamura,M.Yao,I.Tanaka
|
Key ref:

|
 |
A.Nakamura
et al.
(2006).
Ammonia channel couples glutaminase with transamidase reactions in GatCAB.
Science,
312,
1954-1958.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
23-Feb-06
|
Release date:
|
18-Jul-06
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P63488
(GATA_STAAM) -
Glutamyl-tRNA(Gln) amidotransferase subunit A from Staphylococcus aureus (strain Mu50 / ATCC 700699)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
485 a.a.
485 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
 |
|
|
 |
 |
 |
 |
Enzyme class 2:
|
 |
Chain A:
E.C.6.3.5.7
- glutaminyl-tRNA synthase (glutamine-hydrolyzing).
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
L-glutamyl-tRNA(Gln) + L-glutamine + ATP + H2O = L-glutaminyl-tRNA(Gln) + L-glutamate + ADP + phosphate + H+
|
 |
 |
 |
 |
 |
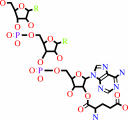 L-glutamyl-tRNA(Gln)
L-glutamyl-tRNA(Gln)
|
+
|
 L-glutamine
L-glutamine
|
+
|
 ATP
ATP
|
+
|
H2O
|
=
|
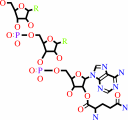 L-glutaminyl-tRNA(Gln)
L-glutaminyl-tRNA(Gln)
|
+
|
 L-glutamate
L-glutamate
|
+
|
 ADP
ADP
|
+
|
 phosphate
phosphate
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 3:
|
 |
Chains B, C:
E.C.6.3.5.-
- ?????
|
|
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Science
312:1954-1958
(2006)
|
|
PubMed id:
|
|
|
|

|
| |
|
Ammonia channel couples glutaminase with transamidase reactions in GatCAB.
|
|
A.Nakamura,
M.Yao,
S.Chimnaronk,
N.Sakai,
I.Tanaka.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The formation of glutaminyl transfer RNA (Gln-tRNA(Gln)) differs among the three
domains of life. Most bacteria employ an indirect pathway to produce
Gln-tRNA(Gln) by a heterotrimeric glutamine amidotransferase CAB (GatCAB) that
acts on the misacylated Glu-tRNA(Gln). Here, we describe a series of crystal
structures of intact GatCAB from Staphylococcus aureus in the apo form and in
the complexes with glutamine, asparagine, Mn2+, and adenosine triphosphate
analog. Two identified catalytic centers for the glutaminase and transamidase
reactions are markedly distant but connected by a hydrophilic ammonia channel 30
A in length. Further, we show that the first U-A base pair in the acceptor stem
and the D loop of tRNA(Gln) serve as identity elements essential for
discrimination by GatCAB and propose a complete model for the overall concerted
reactions to synthesize Gln-tRNA(Gln).

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
Fig. 1. Bacterial GatCAB complex fastens a molecular belt. (A)
Front-view ribbon diagram of the overall structure of S. aureus
GatCAB/glutamine complex at 2.3 Å resolution, depicted in
three different colors for each subunit: blue, green, and
magenta for GatA, GatB, and GatC, respectively. Glutamine in the
active site of GatA is drawn as yellow stick representations,
whereas the purple sphere is the magnesium ion found in the
active site of GatB. ADP from the cocrystal structure with
ADP-AlF[4]^- is shown together as pink sticks. This color code
is used throughout all the figures. (B) Top view of annularly
shaped GatC. (C) Amphipathic helices at the N terminus of GatC
form a helical bundle with the hydrophobic core of GatA
(hydrophobic residues are colored gray). (D) Detailed
interactions between the internal loop region of GatC and GatAB
complex. Conserved residues involved are labeled, with hydrogen
bonds indicated (  3.2 Å, dashed
black line). Polar interactions are prominent on the C-terminal
side, whereas the hydrophobic interactions are clustered on the
opposite side. (E) The C terminus of GatC tightens GatAB
complex, constructing an antiparallel ß sheet and a
hydrophobic platform. 3.2 Å, dashed
black line). Polar interactions are prominent on the C-terminal
side, whereas the hydrophobic interactions are clustered on the
opposite side. (E) The C terminus of GatC tightens GatAB
complex, constructing an antiparallel ß sheet and a
hydrophobic platform.
|
 |
Figure 2.
Fig. 2. A 30 Å long ammonia channel connects the two
remote active centers of GatCAB. (A) The active site of a
glutaminase reaction in GatA is composed of the conserved
Ser-cis-Ser-Lys catalytic scissors shown as magenta stick
representations. Residues involved in the hydrogen-bonded
network (dashed black lines) in the active site are labeled. A
plausible hydrolytic water molecule is colored light blue and is
on the opposite side of a supposed ammonia product (orange
sphere). The Fo-Fc electron density map (contoured at 3  ,
green mesh) calculated without the glutamine and Ser178 clearly
demonstrates the tetrahedral covalent intermediate of the
glutamine with Ser178. (B) The environment of the ADP binding
site shown together with the omit Fo-Fc electron density map (2 ,
green mesh) calculated without the glutamine and Ser178 clearly
demonstrates the tetrahedral covalent intermediate of the
glutamine with Ser178. (B) The environment of the ADP binding
site shown together with the omit Fo-Fc electron density map (2
 , blue). Residues
contributing to ADP (ball-and-stick) recognition are represented
as stick models with labels. Two water molecules (light blue
spheres) are coordinated to a magnesium ion (purple) and to
ß phosphate. (C) The putative ammonia channel was
calculated using the program CAVER (26), with the structure of
the water-omitted GatCAB/glutamine complex. Glu125B blocking the
ammonia transport route is shown in a space-filling
representation for clarity. The channel was filled with a row of
solvent molecules (light blue spheres), which interact with the
conserved polar residues (colored sticks) along the pathway. A
bound glutamine in GatA is drawn as spheres indicating the start
point of the channel. (D) Schematic representation of the
ammonia channel. Residues defining the channel are colored
corresponding to their properties: red, negative; blue,
positive; black, nonpolar side chain; gray, main chain.
Hydrolyzed ammonia is colored orange. Strictly conserved
residues are underlined and hydrogen-bonding distances are
indicated (Å). The presumed movement of the Glu125B gate
to open the ammonia channel is indicated by a black arrow. , blue). Residues
contributing to ADP (ball-and-stick) recognition are represented
as stick models with labels. Two water molecules (light blue
spheres) are coordinated to a magnesium ion (purple) and to
ß phosphate. (C) The putative ammonia channel was
calculated using the program CAVER (26), with the structure of
the water-omitted GatCAB/glutamine complex. Glu125B blocking the
ammonia transport route is shown in a space-filling
representation for clarity. The channel was filled with a row of
solvent molecules (light blue spheres), which interact with the
conserved polar residues (colored sticks) along the pathway. A
bound glutamine in GatA is drawn as spheres indicating the start
point of the channel. (D) Schematic representation of the
ammonia channel. Residues defining the channel are colored
corresponding to their properties: red, negative; blue,
positive; black, nonpolar side chain; gray, main chain.
Hydrolyzed ammonia is colored orange. Strictly conserved
residues are underlined and hydrogen-bonding distances are
indicated (Å). The presumed movement of the Glu125B gate
to open the ammonia channel is indicated by a black arrow.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the AAAs:
Science
(2006,
312,
1954-1958)
copyright 2006.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
A.Nakamura,
K.Sheppard,
J.Yamane,
M.Yao,
D.Söll,
and
I.Tanaka
(2010).
Two distinct regions in Staphylococcus aureus GatCAB guarantee accurate tRNA recognition.
|
| |
Nucleic Acids Res,
38,
672-682.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
K.Yasuhira,
N.Shibata,
G.Mongami,
Y.Uedo,
Y.Atsumi,
Y.Kawashima,
A.Hibino,
Y.Tanaka,
Y.H.Lee,
D.Kato,
M.Takeo,
Y.Higuchi,
and
S.Negoro
(2010).
X-ray crystallographic analysis of the 6-aminohexanoate cyclic dimer hydrolase: catalytic mechanism and evolution of an enzyme responsible for nylon-6 byproduct degradation.
|
| |
J Biol Chem,
285,
1239-1248.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.Blaise,
M.Bailly,
M.Frechin,
M.A.Behrens,
F.Fischer,
C.L.Oliveira,
H.D.Becker,
J.S.Pedersen,
S.Thirup,
and
D.Kern
(2010).
Crystal structure of a transfer-ribonucleoprotein particle that promotes asparagine formation.
|
| |
EMBO J,
29,
3118-3129.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
R.Banerjee,
S.Chen,
K.Dare,
M.Gilreath,
M.Praetorius-Ibba,
M.Raina,
N.M.Reynolds,
T.Rogers,
H.Roy,
S.S.Yadavalli,
and
M.Ibba
(2010).
tRNAs: cellular barcodes for amino acids.
|
| |
FEBS Lett,
584,
387-395.
|
 |
|
|
|
|
 |
T.Ito,
N.Kiyasu,
R.Matsunaga,
S.Takahashi,
and
S.Yokoyama
(2010).
Structure of nondiscriminating glutamyl-tRNA synthetase from Thermotoga maritima.
|
| |
Acta Crystallogr D Biol Crystallogr,
66,
813-820.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
T.Ito,
and
S.Yokoyama
(2010).
Two enzymes bound to one transfer RNA assume alternative conformations for consecutive reactions.
|
| |
Nature,
467,
612-616.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
A.Nagao,
T.Suzuki,
T.Katoh,
Y.Sakaguchi,
and
T.Suzuki
(2009).
Biogenesis of glutaminyl-mt tRNAGln in human mitochondria.
|
| |
Proc Natl Acad Sci U S A,
106,
16209-16214.
|
 |
|
|
|
|
 |
J.Wu,
W.Bu,
K.Sheppard,
M.Kitabatake,
S.T.Kwon,
D.Söll,
and
J.L.Smith
(2009).
Insights into tRNA-dependent amidotransferase evolution and catalysis from the structure of the Aquifex aeolicus enzyme.
|
| |
J Mol Biol,
391,
703-716.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
K.M.Chang,
and
T.L.Hendrickson
(2009).
Recognition of tRNAGln by Helicobacter pylori GluRS2--a tRNAGln-specific glutamyl-tRNA synthetase.
|
| |
Nucleic Acids Res,
37,
6942-6949.
|
 |
|
|
|
|
 |
N.LaRonde-LeBlanc,
M.Resto,
and
B.Gerratana
(2009).
Regulation of active site coupling in glutamine-dependent NAD(+) synthetase.
|
| |
Nat Struct Mol Biol,
16,
421-429.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
P.F.Wang,
A.Yep,
G.L.Kenyon,
and
M.J.McLeish
(2009).
Using directed evolution to probe the substrate specificity of mandelamide hydrolase.
|
| |
Protein Eng Des Sel,
22,
103-110.
|
 |
|
|
|
|
 |
S.Chimnaronk,
F.Forouhar,
J.Sakai,
M.Yao,
C.M.Tron,
M.Atta,
M.Fontecave,
J.F.Hunt,
and
I.Tanaka
(2009).
Snapshots of dynamics in synthesizing N(6)-isopentenyladenosine at the tRNA anticodon.
|
| |
Biochemistry,
48,
5057-5065.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
S.Chimnaronk,
T.Suzuki,
T.Manita,
Y.Ikeuchi,
M.Yao,
T.Suzuki,
and
I.Tanaka
(2009).
RNA helicase module in an acetyltransferase that modifies a specific tRNA anticodon.
|
| |
EMBO J,
28,
1362-1373.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
S.Panjikar,
V.Parthasarathy,
V.S.Lamzin,
M.S.Weiss,
and
P.A.Tucker
(2009).
On the combination of molecular replacement and single-wavelength anomalous diffraction phasing for automated structure determination.
|
| |
Acta Crystallogr D Biol Crystallogr,
65,
1089-1097.
|
 |
|
|
|
|
 |
W.W.Han,
Y.Wang,
Y.H.Zhou,
Y.Yao,
Z.S.Li,
and
Y.Feng
(2009).
Understanding structural/functional properties of amidase from Rhodococcus erythropolis by computational approaches.
|
| |
J Mol Model,
15,
481-487.
|
 |
|
|
|
|
 |
Y.Araiso,
R.L.Sherrer,
R.Ishitani,
J.M.Ho,
D.Söll,
and
O.Nureki
(2009).
Structure of a tRNA-dependent kinase essential for selenocysteine decoding.
|
| |
Proc Natl Acad Sci U S A,
106,
16215-16220.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
Y.Tanaka,
S.Yamagata,
Y.Kitago,
Y.Yamada,
S.Chimnaronk,
M.Yao,
and
I.Tanaka
(2009).
Deduced RNA binding mechanism of ThiI based on structural and binding analyses of a minimal RNA ligand.
|
| |
RNA,
15,
1498-1506.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
C.Pujol,
M.Bailly,
D.Kern,
L.Maréchal-Drouard,
H.Becker,
and
A.M.Duchêne
(2008).
Dual-targeted tRNA-dependent amidotransferase ensures both mitochondrial and chloroplastic Gln-tRNAGln synthesis in plants.
|
| |
Proc Natl Acad Sci U S A,
105,
6481-6485.
|
 |
|
|
|
|
 |
J.Yuan,
K.Sheppard,
and
D.Söll
(2008).
Amino acid modifications on tRNA.
|
| |
Acta Biochim Biophys Sin (Shanghai),
40,
539-553.
|
 |
|
|
|
|
 |
K.Sheppard,
and
D.Söll
(2008).
On the evolution of the tRNA-dependent amidotransferases, GatCAB and GatDE.
|
| |
J Mol Biol,
377,
831-844.
|
 |
|
|
|
|
 |
K.Sheppard,
J.Yuan,
M.J.Hohn,
B.Jester,
K.M.Devine,
and
D.Söll
(2008).
From one amino acid to another: tRNA-dependent amino acid biosynthesis.
|
| |
Nucleic Acids Res,
36,
1813-1825.
|
 |
|
|
|
|
 |
K.Sheppard,
R.L.Sherrer,
and
D.Söll
(2008).
Methanothermobacter thermautotrophicus tRNA Gln confines the amidotransferase GatCAB to asparaginyl-tRNA Asn formation.
|
| |
J Mol Biol,
377,
845-853.
|
 |
|
|
|
|
 |
M.A.Vanoni,
and
B.Curti
(2008).
Structure-function studies of glutamate synthases: a class of self-regulated iron-sulfur flavoenzymes essential for nitrogen assimilation.
|
| |
IUBMB Life,
60,
287-300.
|
 |
|
|
|
|
 |
M.Bailly,
M.Blaise,
B.Lorber,
H.D.Becker,
and
D.Kern
(2007).
The transamidosome: a dynamic ribonucleoprotein particle dedicated to prokaryotic tRNA-dependent asparagine biosynthesis.
|
| |
Mol Cell,
28,
228-239.
|
 |
|
|
|
|
 |
M.Deniziak,
C.Sauter,
H.D.Becker,
C.A.Paulus,
R.Giegé,
and
D.Kern
(2007).
Deinococcus glutaminyl-tRNA synthetase is a chimer between proteins from an ancient and the modern pathways of aminoacyl-tRNA formation.
|
| |
Nucleic Acids Res,
35,
1421-1431.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
S.Mouilleron,
and
B.Golinelli-Pimpaneau
(2007).
Conformational changes in ammonia-channeling glutamine amidotransferases.
|
| |
Curr Opin Struct Biol,
17,
653-664.
|
 |
|
|
|
|
 |
S.Namgoong,
K.Sheppard,
R.L.Sherrer,
and
D.Söll
(2007).
Co-evolution of the archaeal tRNA-dependent amidotransferase GatCAB with tRNA(Asn).
|
| |
FEBS Lett,
581,
309-314.
|
 |
|
|
|
|
 |
T.Cathopoulis,
P.Chuawong,
and
T.L.Hendrickson
(2007).
Novel tRNA aminoacylation mechanisms.
|
| |
Mol Biosyst,
3,
408-418.
|
 |
|
|
|
|
 |
M.Bailly,
S.Giannouli,
M.Blaise,
C.Stathopoulos,
D.Kern,
and
H.D.Becker
(2006).
A single tRNA base pair mediates bacterial tRNA-dependent biosynthesis of asparagine.
|
| |
Nucleic Acids Res,
34,
6083-6094.
|
 |
|
|
|
|
 |
M.J.Hohn,
H.S.Park,
P.O'Donoghue,
M.Schnitzbauer,
and
D.Söll
(2006).
Emergence of the universal genetic code imprinted in an RNA record.
|
| |
Proc Natl Acad Sci U S A,
103,
18095-18100.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |
|

