 |
PDBsum entry 3al0

|

|

|
|
 |
Contents |
 |
|
|

|

|

|

|

|

|

|
 472 a.a.
472 a.a.
|
 |

|

|

|

|

|

|

|
 482 a.a.
482 a.a.
|
 |

|

|

|

|

|

|

|
 564 a.a.
564 a.a.
|
 |

|

|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Ligase/RNA
|
 |
|
Title:
|
 |
Crystal structure of the glutamine transamidosome from thermotoga maritima in the glutamylation state.
|
|
Structure:
|
 |
Glutamyl-tRNA(gln) amidotransferase subunit a. Chain: a. Synonym: glu-adt subunit a. Engineered: yes. Aspartyl/glutamyl-tRNA(asn/gln) amidotransferase subunit b. Chain: b. Synonym: asp/glu-adt subunit b. Engineered: yes. Glutamyl-tRNA(gln) amidotransferase subunit c,linker,
|
|
Source:
|
 |
Thermotoga maritima. Organism_taxid: 243274. Strain: atcc 43589 / msb8 / dsm 3109 / jcm 10099. Gene: gata, tm_1272. Expressed in: escherichia coli. Expression_system_taxid: 562. Gene: gatb, tm_1273. Thermotoga maritima, synthetic construct. Organism_taxid: 243274, 32630.
|
|
Resolution:
|
 |
|
3.37Å
|
R-factor:
|
0.199
|
R-free:
|
0.269
|
|
|
Authors:
|
 |
T.Ito,S.Yokoyama
|
|
Key ref:
|
 |
T.Ito
and
S.Yokoyama
(2010).
Two enzymes bound to one transfer RNA assume alternative conformations for consecutive reactions.
Nature,
467,
612-616.
PubMed id:
|
 |
|
Date:
|
 |
|
19-Jul-10
|
Release date:
|
29-Sep-10
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
Q9X0Z9
(GATA_THEMA) -
Glutamyl-tRNA(Gln) amidotransferase subunit A from Thermotoga maritima (strain ATCC 43589 / DSM 3109 / JCM 10099 / NBRC 100826 / MSB8)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
475 a.a.
472 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|

|

|
|
Q9X100
(GATB_THEMA) -
Aspartyl/glutamyl-tRNA(Asn/Gln) amidotransferase subunit B from Thermotoga maritima (strain ATCC 43589 / DSM 3109 / JCM 10099 / NBRC 100826 / MSB8)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
482 a.a.
482 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
 |
|
|
 |
 |
 |
 |
Enzyme class 2:
|
 |
Chain A:
E.C.6.3.5.7
- glutaminyl-tRNA synthase (glutamine-hydrolyzing).
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
L-glutamyl-tRNA(Gln) + L-glutamine + ATP + H2O = L-glutaminyl-tRNA(Gln) + L-glutamate + ADP + phosphate + H+
|
 |
 |
 |
 |
 |
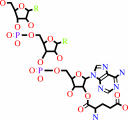 L-glutamyl-tRNA(Gln)
L-glutamyl-tRNA(Gln)
|
+
|
 L-glutamine
L-glutamine
|
+
|
 ATP
ATP
|
+
|
H2O
|
=
|
L-glutaminyl-tRNA(Gln)
Bound ligand (Het Group name = )
matches with 47.50% similarity
|
+
|
 L-glutamate
L-glutamate
|
+
|
 ADP
ADP
|
+
|
 phosphate
phosphate
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 3:
|
 |
Chains B, C:
E.C.6.3.5.-
- ?????
|
|
 |
 |
 |
 |
 |
Enzyme class 4:
|
 |
Chain C:
E.C.6.1.1.17
- glutamate--tRNA ligase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
tRNA(Glu) + L-glutamate + ATP = L-glutamyl-tRNA(Glu) + AMP + diphosphate
|
 |
 |
 |
 |
 |
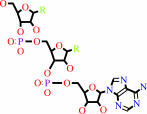 tRNA(Glu)
tRNA(Glu)
|
+
|
 L-glutamate
L-glutamate
|
+
|
 ATP
ATP
|
=
|
L-glutamyl-tRNA(Glu)
Bound ligand (Het Group name = )
matches with 52.78% similarity
|
+
|
 AMP
AMP
|
+
|
 diphosphate
diphosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
Nature
467:612-616
(2010)
|
|
PubMed id:
|
|
|
|

|
| |
|
Two enzymes bound to one transfer RNA assume alternative conformations for consecutive reactions.
|
|
T.Ito,
S.Yokoyama.
|
|
|

|
| |
ABSTRACT
|
|

|
| |
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
F.J.Blanco,
and
G.Montoya
(2011).
Transient DNA / RNA-protein interactions.
|
| |
FEBS J,
278,
1643-1650.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
|

| | |

