 |
PDBsum entry 1vbm

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Ligase
|
 |
|
Title:
|
 |
Crystal structure of the escherichia coli tyrosyl-tRNA synthetase complexed with tyr-ams
|
|
Structure:
|
 |
Tyrosyl-tRNA synthetase. Chain: a, b. Fragment: residues 5-322. Synonym: tyrosine--tRNA ligase, tyrrs. Engineered: yes
|
|
Source:
|
 |
Escherichia coli. Organism_taxid: 562. Expressed in: escherichia coli bl21(de3). Expression_system_taxid: 469008.
|
|
Biol. unit:
|
 |
Dimer (from
)
|
|
Resolution:
|
 |
|
2.70Å
|
R-factor:
|
0.246
|
R-free:
|
0.284
|
|
|
Authors:
|
 |
T.Kobayashi,K.Sakamoto,T.Takimura,K.Kamata,R.Sekine,S.Nishimura, S.Yokoyama,Riken Structural Genomics/proteomics Initiative (Rsgi)
|
Key ref:

|
 |
T.Kobayashi
et al.
(2005).
Structural snapshots of the KMSKS loop rearrangement for amino acid activation by bacterial tyrosyl-tRNA synthetase.
J Mol Biol,
346,
105-117.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
27-Feb-04
|
Release date:
|
25-Jan-05
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P0AGJ9
(SYY_ECOLI) -
Tyrosine--tRNA ligase from Escherichia coli (strain K12)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
424 a.a.
318 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.6.1.1.1
- tyrosine--tRNA ligase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
tRNA(Tyr) + L-tyrosine + ATP = L-tyrosyl-tRNA(Tyr) + AMP + diphosphate + H+
|
 |
 |
 |
 |
 |
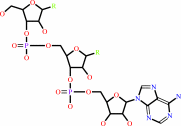 tRNA(Tyr)
tRNA(Tyr)
|
+
|
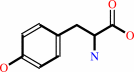 L-tyrosine
L-tyrosine
|
+
|
 ATP
ATP
|
=
|
L-tyrosyl-tRNA(Tyr)
Bound ligand (Het Group name = )
matches with 48.72% similarity
|
+
|
 AMP
AMP
|
+
|
 diphosphate
diphosphate
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Mol Biol
346:105-117
(2005)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structural snapshots of the KMSKS loop rearrangement for amino acid activation by bacterial tyrosyl-tRNA synthetase.
|
|
T.Kobayashi,
T.Takimura,
R.Sekine,
V.P.Kelly,
K.Vincent,
K.Kamata,
K.Sakamoto,
S.Nishimura,
S.Yokoyama.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Tyrosyl-tRNA synthetase (TyrRS) has been studied extensively by mutational and
structural analyses to elucidate its catalytic mechanism. TyrRS has the HIGH and
KMSKS motifs that catalyze the amino acid activation with ATP. In the present
study, the crystal structures of the Escherichia coli TyrRS catalytic domain, in
complexes with l-tyrosine and a l-tyrosyladenylate analogue, Tyr-AMS, were
solved at 2.0A and 2.7A resolution, respectively. In the Tyr-AMS-bound
structure, the 2'-OH group and adenine ring of the Tyr-AMS are strictly
recognized by hydrogen bonds. This manner of hydrogen-bond recognition is
conserved among the class I synthetases. Moreover, a comparison between the two
structures revealed that the KMSKS loop is rearranged in response to adenine
moiety binding and hydrogen-bond formation, and the KMSKS loop adopts the more
compact ("semi-open") form, rather than the flexible, open form. The
HIGH motif initially recognizes the gamma-phosphate, and then the alpha and
gamma-phosphates of ATP, with a slight rearrangement of the residues. The other
residues around the substrate also accommodate the Tyr-AMS. This induced-fit
form presents a novel "snapshot" of the amino acid activation step in
the aminoacylation reaction by TyrRS. The present structures and the
T.thermophilus TyrRS ATP-free and bound structures revealed that the extensive
induced-fit conformational changes of the KMSKS loop and the local
conformational changes within the substrate binding site form the basis for
driving the amino acid activation step: the KMSKS loop adopts the open form,
transiently shifts to the semi-open conformation according to the adenosyl
moiety binding, and finally assumes the rigid ATP-bound, closed form. After the
amino acid activation, the KMSKS loop adopts the semi-open form again to accept
the CCA end of tRNA for the aminoacyl transfer reaction.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 6.
Figure 6. The Tyr-AMP binding sites of the TyrRSs. (a) The
Tyr-AMP-binding site of the E. coli TyrRS·Tyr-AMS
complex. (b) The corresponding view of the B. stearothermophilus
TyrRS·Tyr-AMP complex5 (PDB ID: 3TS1). The carbon atoms
of Tyr-AMS/Tyr-AMP are shown in light blue, and the hydrogen
bonds are indicated by green broken lines. A phosphate atom is
shown in green.
|
 |
Figure 8.
Figure 8. Conformational changes of the KMSKS loop. (a)
Open conformation of the KMSKS loop in the E. coli
TyrRS·L-tyrosine structure. (b) Semi-open conformation of
the KMSKS loop in the E. coil TyrRS·Tyr-AMS structure.
The superposition of the open-form KMSKS loop is also shown by a
pink translucent tube. (c) Closed conformation of the KMSKS loop
in the T. thermophilus
TyrRS·ATP·L-tyrosinol·tRNA^Tyr structure13
(PDB ID: 1H3E). The KMSKS loops are shown by orange tubes. The
carbon atoms of L-tyrosine and L-tyrosinol are shown in pink,
and those of Tyr-AMS are shown in light blue, respectively. (d)
The bottleneck of the catalytic site in the T. thermophilus
TyrRS closed form. The surface model of Tyr-AMP phosphate, which
is a landmark of the aminoacyl transfer center, is shown by a
stick model. (e) The 3'-adenosine of the tRNA cannot pass
through the bottleneck of (d). The adenosine moiety is shown by
a CPK model. (f) The exposed catalytic site in the E. coli TyrRS
semi-open form. Tyr-AMS is shown by a stick model. The molecular
surfaces were produced using the program MSMS
(http://www.scripps.edu/mb/olson/people/sanner/html/msms_home.html).
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from Elsevier:
J Mol Biol
(2005,
346,
105-117)
copyright 2005.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
A.Palencia,
T.Crépin,
M.T.Vu,
T.L.Lincecum,
S.A.Martinis,
and
S.Cusack
(2012).
Structural dynamics of the aminoacylation and proofreading functional cycle of bacterial leucyl-tRNA synthetase.
|
| |
Nat Struct Mol Biol,
19,
677-684.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
I.T.Yonemoto,
and
E.M.Tippmann
(2010).
The juggernauts of biology.
|
| |
Bioessays,
32,
314-321.
|
 |
|
|
|
|
 |
M.Zhou,
X.Dong,
N.Shen,
C.Zhong,
and
J.Ding
(2010).
Crystal structures of Saccharomyces cerevisiae tryptophanyl-tRNA synthetase: new insights into the mechanism of tryptophan activation and implications for anti-fungal drug design.
|
| |
Nucleic Acids Res,
38,
3399-3413.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
X.Dong,
M.Zhou,
C.Zhong,
B.Yang,
N.Shen,
and
J.Ding
(2010).
Crystal structure of Pyrococcus horikoshii tryptophanyl-tRNA synthetase and structure-based phylogenetic analysis suggest an archaeal origin of tryptophanyl-tRNA synthetase.
|
| |
Nucleic Acids Res,
38,
1401-1412.
|
 |
|
|
|
|
 |
X.L.Zhou,
M.Wang,
M.Tan,
Q.Huang,
G.Eriani,
and
E.D.Wang
(2010).
Functional characterization of leucine-specific domain 1 from eukaryal and archaeal leucyl-tRNA synthetases.
|
| |
Biochem J,
429,
505-513.
|
 |
|
|
|
|
 |
G.Sharma,
and
E.A.First
(2009).
Thermodynamic Analysis Reveals a Temperature-dependent Change in the Catalytic Mechanism of Bacillus stearothermophilus Tyrosyl-tRNA Synthetase.
|
| |
J Biol Chem,
284,
4179-4190.
|
 |
|
|
|
|
 |
J.K.Takimoto,
K.L.Adams,
Z.Xiang,
and
L.Wang
(2009).
Improving orthogonal tRNA-synthetase recognition for efficient unnatural amino acid incorporation and application in mammalian cells.
|
| |
Mol Biosyst,
5,
931-934.
|
 |
|
|
|
|
 |
J.Lee,
J.Johnson,
Z.Ding,
M.Paetzel,
and
R.B.Cornell
(2009).
Crystal structure of a mammalian CTP: phosphocholine cytidylyltransferase catalytic domain reveals novel active site residues within a highly conserved nucleotidyltransferase fold.
|
| |
J Biol Chem,
284,
33535-33548.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
K.Sakamoto,
K.Murayama,
K.Oki,
F.Iraha,
M.Kato-Murayama,
M.Takahashi,
K.Ohtake,
T.Kobayashi,
S.Kuramitsu,
M.Shirouzu,
and
S.Yokoyama
(2009).
Genetic encoding of 3-iodo-L-tyrosine in Escherichia coli for single-wavelength anomalous dispersion phasing in protein crystallography.
|
| |
Structure,
17,
335-344.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
S.Kamijo,
A.Fujii,
K.Onodera,
and
K.Wakabayashi
(2009).
Analyses of conditions for KMSSS loop in tyrosyl-tRNA synthetase by building a mutant library.
|
| |
J Biochem,
146,
241-250.
|
 |
|
|
|
|
 |
N.Shen,
M.Zhou,
B.Yang,
Y.Yu,
X.Dong,
and
J.Ding
(2008).
Catalytic mechanism of the tryptophan activation reaction revealed by crystal structures of human tryptophanyl-tRNA synthetase in different enzymatic states.
|
| |
Nucleic Acids Res,
36,
1288-1299.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
T.L.Hendrickson
(2008).
Proofreading optimizes iodotyrosine insertion into the genetic code.
|
| |
Proc Natl Acad Sci U S A,
105,
13699-13700.
|
 |
|
|
|
|
 |
T.Li,
M.Froeyen,
and
P.Herdewijn
(2008).
Comparative structural dynamics of Tyrosyl-tRNA synthetase complexed with different substrates explored by molecular dynamics.
|
| |
Eur Biophys J,
38,
25-35.
|
 |
|
|
|
|
 |
D.Thompson,
C.Lazennec,
P.Plateau,
and
T.Simonson
(2007).
Ammonium scanning in an enzyme active site. The chiral specificity of aspartyl-tRNA synthetase.
|
| |
J Biol Chem,
282,
30856-30868.
|
 |
|
|
|
|
 |
I.Kufareva,
L.Budagyan,
E.Raush,
M.Totrov,
and
R.Abagyan
(2007).
PIER: protein interface recognition for structural proteomics.
|
| |
Proteins,
67,
400-417.
|
 |
|
|
|
|
 |
L.Bonnefond,
M.Frugier,
E.Touzé,
B.Lorber,
C.Florentz,
R.Giegé,
J.Rudinger-Thirion,
and
C.Sauter
(2007).
Tyrosyl-tRNA synthetase: the first crystallization of a human mitochondrial aminoacyl-tRNA synthetase.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
63,
338-341.
|
 |
|
|
|
|
 |
M.E.Budiman,
M.H.Knaggs,
J.S.Fetrow,
and
R.W.Alexander
(2007).
Using molecular dynamics to map interaction networks in an aminoacyl-tRNA synthetase.
|
| |
Proteins,
68,
670-689.
|
 |
|
|
|
|
 |
M.T.Vu,
and
S.A.Martinis
(2007).
A unique insert of leucyl-tRNA synthetase is required for aminoacylation and not amino acid editing.
|
| |
Biochemistry,
46,
5170-5176.
|
 |
|
|
|
|
 |
M.Tsunoda,
Y.Kusakabe,
N.Tanaka,
S.Ohno,
M.Nakamura,
T.Senda,
T.Moriguchi,
N.Asai,
M.Sekine,
T.Yokogawa,
K.Nishikawa,
and
K.T.Nakamura
(2007).
Structural basis for recognition of cognate tRNA by tyrosyl-tRNA synthetase from three kingdoms.
|
| |
Nucleic Acids Res,
35,
4289-4300.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
T.A.Cropp,
J.C.Anderson,
and
J.W.Chin
(2007).
Reprogramming the amino-acid substrate specificity of orthogonal aminoacyl-tRNA synthetases to expand the genetic code of eukaryotic cells.
|
| |
Nat Protoc,
2,
2590-2600.
|
 |
|
|
|
|
 |
L.Wang,
J.Xie,
and
P.G.Schultz
(2006).
Expanding the genetic code.
|
| |
Annu Rev Biophys Biomol Struct,
35,
225-249.
|
 |
|
|
|
|
 |
T.Kobayashi,
K.Sakamoto,
T.Takimura,
R.Sekine,
V.P.Kelly,
K.Vincent,
K.Kamata,
S.Nishimura,
and
S.Yokoyama
(2005).
Structural basis of nonnatural amino acid recognition by an engineered aminoacyl-tRNA synthetase for genetic code expansion.
|
| |
Proc Natl Acad Sci U S A,
102,
1366-1371.
|
 |
|
PDB codes:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

