 |
PDBsum entry 1s2a

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
1s2a
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Oxidoreductase
|
 |
|
Title:
|
 |
Crystal structures of prostaglandin d2 11-ketoreductase in complex with the non-steroidal anti-inflammatory drugs flufenamic acid and indomethacin
|
|
Structure:
|
 |
Aldo-keto reductase family 1 member c3. Chain: a. Synonym: akr1c3, prostaglandin d2 11-ketoreductase, prostaglandin f synthase, 3-alpha-hsd-type-2, 17-beta-hsd-type-5. Engineered: yes
|
|
Source:
|
 |
Homo sapiens. Human. Organism_taxid: 9606. Expressed in: escherichia coli. Expression_system_taxid: 562.
|
|
Resolution:
|
 |
|
1.70Å
|
R-factor:
|
0.174
|
R-free:
|
0.198
|
|
|
Authors:
|
 |
A.L.Lovering,J.P.Ride,C.M.Bunce,J.C.Desmond,S.M.Cummings,S.A.White
|
|
Key ref:
|
 |
A.L.Lovering
et al.
(2004).
Crystal structures of prostaglandin D(2) 11-ketoreductase (AKR1C3) in complex with the nonsteroidal anti-inflammatory drugs flufenamic acid and indomethacin.
Cancer Res,
64,
1802-1810.
PubMed id:
DOI:
|
 |
|
Date:
|
 |
|
08-Jan-04
|
Release date:
|
23-Mar-04
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P42330
(AK1C3_HUMAN) -
Aldo-keto reductase family 1 member C3 from Homo sapiens
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
323 a.a.
315 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 1:
|
 |
E.C.1.1.1.188
- prostaglandin-F synthase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
prostaglandin F2alpha + NADP+ = prostaglandin D2 + NADPH + H+
|
 |
 |
 |
 |
 |
prostaglandin F2alpha
Bound ligand (Het Group name = )
corresponds exactly
|
+
|
 NADP(+)
NADP(+)
|
=
|
 prostaglandin D2
prostaglandin D2
|
+
|
 NADPH
NADPH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.1.1.1.210
- 3beta-(or 20alpha)-hydroxysteroid dehydrogenase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
5alpha-androstane-3beta,17beta-diol + NADP+ = 17beta-hydroxy-5alpha- androstan-3-one + NADPH + H+
|
 |
 |
 |
 |
 |
5alpha-androstane-3beta,17beta-diol
|
+
|
NADP(+)
Bound ligand (Het Group name = )
corresponds exactly
|
=
|
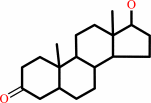 17beta-hydroxy-5alpha- androstan-3-one
17beta-hydroxy-5alpha- androstan-3-one
|
+
|
 NADPH
NADPH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 3:
|
 |
E.C.1.1.1.239
- 3alpha-(17beta)-hydroxysteroid dehydrogenase (NAD(+)).
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
testosterone + NAD+ = androst-4-ene-3,17-dione + NADH + H+
|
 |
 |
 |
 |
 |
testosterone
Bound ligand (Het Group name = )
matches with 91.67% similarity
|
+
|
 NAD(+)
NAD(+)
|
=
|
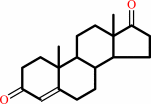 androst-4-ene-3,17-dione
androst-4-ene-3,17-dione
|
+
|
 NADH
NADH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 4:
|
 |
E.C.1.1.1.357
- 3alpha-hydroxysteroid 3-dehydrogenase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
|
1.
|
a 3alpha-hydroxysteroid + NADP+ = a 3-oxosteroid + NADPH + H+
|
|
2.
|
a 3alpha-hydroxysteroid + NAD+ = a 3-oxosteroid + NADH + H+
|
|
 |
 |
 |
 |
 |
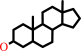 3alpha-hydroxysteroid
3alpha-hydroxysteroid
|
+
|
NADP(+)
Bound ligand (Het Group name = )
corresponds exactly
|
=
|
 3-oxosteroid
3-oxosteroid
|
+
|
 NADPH
NADPH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 3alpha-hydroxysteroid
3alpha-hydroxysteroid
|
+
|
NAD(+)
Bound ligand (Het Group name = )
matches with 91.67% similarity
|
=
|
 3-oxosteroid
3-oxosteroid
|
+
|
 NADH
NADH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 5:
|
 |
E.C.1.1.1.53
- 3alpha(or 20beta)-hydroxysteroid dehydrogenase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
androstan-3alpha,17beta-diol + NAD+ = 17beta-hydroxyandrostanone + NADH + H+
|
 |
 |
 |
 |
 |
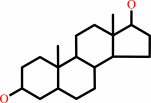 androstan-3alpha,17beta-diol
androstan-3alpha,17beta-diol
|
+
|
NAD(+)
Bound ligand (Het Group name = )
matches with 91.67% similarity
|
=
|
17beta-hydroxyandrostanone
|
+
|
 NADH
NADH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 6:
|
 |
E.C.1.1.1.62
- 17beta-estradiol 17-dehydrogenase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
|
1.
|
17beta-estradiol + NAD+ = estrone + NADH + H+
|
|
2.
|
17beta-estradiol + NADP+ = estrone + NADPH + H+
|
|
 |
 |
 |
 |
 |
17beta-estradiol
|
+
|
NAD(+)
Bound ligand (Het Group name = )
matches with 91.67% similarity
|
=
|
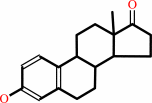 estrone
estrone
|
+
|
 NADH
NADH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
17beta-estradiol
|
+
|
NADP(+)
Bound ligand (Het Group name = )
corresponds exactly
|
=
|
 estrone
estrone
|
+
|
 NADPH
NADPH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 7:
|
 |
E.C.1.1.1.64
- testosterone 17beta-dehydrogenase (NADP(+)).
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
testosterone + NADP+ = androst-4-ene-3,17-dione + NADPH + H+
|
 |
 |
 |
 |
 |
testosterone
Bound ligand (Het Group name = )
corresponds exactly
|
+
|
 NADP(+)
NADP(+)
|
=
|
 androst-4-ene-3,17-dione
androst-4-ene-3,17-dione
|
+
|
 NADPH
NADPH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Cancer Res
64:1802-1810
(2004)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structures of prostaglandin D(2) 11-ketoreductase (AKR1C3) in complex with the nonsteroidal anti-inflammatory drugs flufenamic acid and indomethacin.
|
|
A.L.Lovering,
J.P.Ride,
C.M.Bunce,
J.C.Desmond,
S.M.Cummings,
S.A.White.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
It is becoming increasingly well established that nonsteroidal anti-inflammatory
drugs (NSAID) protect against tumors of the gastrointestinal tract and that they
may also protect against a variety of other tumors. These activities have been
widely attributed to the inhibition of cylooxygenases (COX) and, in particular,
COX-2. However, several observations have indicated that other targets may be
involved. Besides targeting COX, certain NSAID also inhibit enzymes belonging to
the aldo-keto reductase (AKR) family, including AKR1C3. We have demonstrated
previously that overexpression of AKR1C3 acts to suppress cell differentiation
and promote proliferation in myeloid cells. However, this enzyme has a broad
tissue distribution and therefore represents a novel candidate for the target of
the COX-independent antineoplastic actions of NSAID. Here we report on the X-ray
crystal structures of AKR1C3 complexed with the NSAID indomethacin (1.8 A
resolution) or flufenamic acid (1.7 A resolution). One molecule of indomethacin
is bound in the active site, whereas flufenamic acid binds to both the active
site and the beta-hairpin loop, at the opposite end of the central beta-barrel.
Two other crystal structures (1.20 and 2.1 A resolution) show acetate bound in
the active site occupying the proposed oxyanion hole. The data underline AKR1C3
as a COX-independent target for NSAID and will provide a structural basis for
the future development of new cancer therapies with reduced COX-dependent side
effects.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
F.L.Khanim,
R.E.Hayden,
J.Birtwistle,
A.Lodi,
S.Tiziani,
N.J.Davies,
J.P.Ride,
M.R.Viant,
U.L.Gunther,
J.C.Mountford,
H.Schrewe,
R.M.Green,
J.A.Murray,
M.T.Drayson,
and
C.M.Bunce
(2009).
Combined bezafibrate and medroxyprogesterone acetate: potential novel therapy for acute myeloid leukaemia.
|
| |
PLoS One,
4,
e8147.
|
 |
|
|
|
|
 |
M.C.Byrns,
and
T.M.Penning
(2009).
Type 5 17beta-hydroxysteroid dehydrogenase/prostaglandin F synthase (AKR1C3): role in breast cancer and inhibition by non-steroidal anti-inflammatory drug analogs.
|
| |
Chem Biol Interact,
178,
221-227.
|
 |
|
|
|
|
 |
P.Veliça,
N.J.Davies,
P.P.Rocha,
H.Schrewe,
J.P.Ride,
and
C.M.Bunce
(2009).
Lack of functional and expression homology between human and mouse aldo-keto reductase 1C enzymes: implications for modelling human cancers.
|
| |
Mol Cancer,
8,
121.
|
 |
|
|
|
|
 |
Y.Jin,
L.Duan,
S.H.Lee,
H.J.Kloosterboer,
I.A.Blair,
and
T.M.Penning
(2009).
Human cytosolic hydroxysteroid dehydrogenases of the aldo-ketoreductase superfamily catalyze reduction of conjugated steroids: implications for phase I and phase II steroid hormone metabolism.
|
| |
J Biol Chem,
284,
10013-10022.
|
 |
|
|
|
|
 |
C.Ludwig,
P.J.Michiels,
A.Lodi,
J.Ride,
C.Bunce,
and
U.L.Günther
(2008).
Evaluation of solvent accessibility epitopes for different dehydrogenase inhibitors.
|
| |
ChemMedChem,
3,
1371-1376.
|
 |
|
|
|
|
 |
M.C.Byrns,
S.Steckelbroeck,
and
T.M.Penning
(2008).
An indomethacin analogue, N-(4-chlorobenzoyl)-melatonin, is a selective inhibitor of aldo-keto reductase 1C3 (type 2 3alpha-HSD, type 5 17beta-HSD, and prostaglandin F synthase), a potential target for the treatment of hormone dependent and hormone independent malignancies.
|
| |
Biochem Pharmacol,
75,
484-493.
|
 |
|
|
|
|
 |
M.J.Campbell,
C.Carlberg,
and
H.P.Koeffler
(2008).
A Role for the PPARgamma in Cancer Therapy.
|
| |
PPAR Res,
2008,
314974.
|
 |
|
|
|
|
 |
Y.H.Wu,
T.P.Ko,
R.T.Guo,
S.M.Hu,
L.M.Chuang,
and
A.H.Wang
(2008).
Structural basis for catalytic and inhibitory mechanisms of human prostaglandin reductase PTGR2.
|
| |
Structure,
16,
1714-1723.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
U.Oppermann
(2007).
Carbonyl reductases: the complex relationships of mammalian carbonyl- and quinone-reducing enzymes and their role in physiology.
|
| |
Annu Rev Pharmacol Toxicol,
47,
293-322.
|
 |
|
|
|
|
 |
T.Matsunaga,
S.Shintani,
and
A.Hara
(2006).
Multiplicity of mammalian reductases for xenobiotic carbonyl compounds.
|
| |
Drug Metab Pharmacokinet,
21,
1.
|
 |
|
|
|
|
 |
J.F.Couture,
K.P.de Jésus-Tran,
A.M.Roy,
L.Cantin,
P.L.Côté,
P.Legrand,
V.Luu-The,
F.Labrie,
and
R.Breton
(2005).
Comparison of crystal structures of human type 3 3alpha-hydroxysteroid dehydrogenase reveals an "induced-fit" mechanism and a conserved basic motif involved in the binding of androgen.
|
| |
Protein Sci,
14,
1485-1497.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
O.El-Kabbani,
S.Ishikura,
A.Wagner,
C.Schulze-Briese,
and
A.Hara
(2005).
Crystallization and preliminary X-ray diffraction analysis of mouse 3(17)alpha-hydroxysteroid dehydrogenase.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
61,
688-690.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

