 |
PDBsum entry 1nm3

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Electron transport
|
PDB id
|
|

|

|
1nm3
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.11.1.27
- glutathione-dependent peroxiredoxin.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
a hydroperoxide + 2 glutathione = an alcohol + glutathione disulfide + H2O
|
 |
 |
 |
 |
 |
 hydroperoxide
hydroperoxide
|
+
|
 2
×
glutathione
2
×
glutathione
|
=
|
 alcohol
alcohol
|
+
|
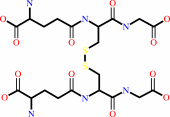 glutathione disulfide
glutathione disulfide
|
+
|
H2O
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Biol Chem
278:10790-10798
(2003)
|
|
PubMed id:
|
|
|
|

|
| |
|
The tetrameric structure of Haemophilus influenza hybrid Prx5 reveals interactions between electron donor and acceptor proteins.
|
|
S.J.Kim,
J.R.Woo,
Y.S.Hwang,
D.G.Jeong,
D.H.Shin,
K.Kim,
S.E.Ryu.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Cellular redox control is often mediated by oxidation and reduction of cysteine
residues in the redox-sensitive proteins, where thioredoxin and glutaredoxin
(Grx) play as electron donors for the oxidized proteins. Despite the importance
of protein-protein interactions between the electron donor and acceptor
proteins, there has been no structural information for the interaction of
thioredoxin or Grx with natural target proteins. Here, we present the crystal
structure of a novel Haemophilus influenza peroxiredoxin (Prx) hybrid Prx5
determined at 2.8-A resolution. The structure reveals that hybrid Prx5 forms a
tightly associated tetramer where active sites of Prx and Grx domains of
different monomers interact with each other. The Prx-Grx interface comprises
specific charge interactions surrounded by weak interactions, providing insight
into the target recognition mechanism of Grx. The tetrameric structure also
exhibits a flexible active site and alternative Prx-Grx interactions, which
appear to facilitate the electron transfer from Grx to Prx domain. Differences
of electron donor binding surfaces in Prx proteins revealed by an analysis based
on the structural information explain the electron donor specificities of
various Prx proteins.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 4.
Fig. 4. Environment of the redox active site. a, Prx-Grx
interaction and glutathione binding in the hyPrx5 tetramer.
Glutathione model (shown in the figure as a ball and stick
diagram) was created by superposing the complex structure of
Grx3-glutathione (PDB code 3GRX) on the hyPrx5 Grx domain.
Surfaces of Prx (monomer A) and Grx (monomer D) domains are
colored red and gold, respectively. Redox active cysteines
(Cys-49 and Cys-180) and residues involved in the Prx-Grx
interaction are labeled. b, stereo view of the 2F[o]  F[c]
electron density map. The electron density map around the Prx
active site of hyPrx5 is presented as superimposed with the
refined model. The map was contoured at a 0.9 F[c]
electron density map. The electron density map around the Prx
active site of hyPrx5 is presented as superimposed with the
refined model. The map was contoured at a 0.9  level. c,
stereo view of the Prx active site of hyPrx5. The side chains of
residues (Arg-126 and Thr-46) contributing to the reactivity of
the sulfur atom of Cys-49 were drawn as a ball-and-stick
representation. Distances between the interacting atoms are
shown in the figure. level. c,
stereo view of the Prx active site of hyPrx5. The side chains of
residues (Arg-126 and Thr-46) contributing to the reactivity of
the sulfur atom of Cys-49 were drawn as a ball-and-stick
representation. Distances between the interacting atoms are
shown in the figure.
|
 |
Figure 5.
Fig. 5. Interaction surfaces implicated in the Prx-Grx or
Prx-Trx interaction. Electrostatic potential surfaces of the
hyPrx5 Prx domain, the hyPrx5 Grx domain, human Prx5 (PDB code
1HD2), and human Trx (PDB code 1ERU) were calculated by using
the program GRASP (27). Positive and negative charges are
represented as blue and red, respectively. a, the surface of the
hyPrx5 Prx domain. Residues involved in the Prx-Grx contact in
the hyPrx5 tetramer are labeled and surrounded by a yellow
lines. The alternative interaction surface (see "Prx-Grx
Interaction") is indicated with a green line. b, the surface of
the hyPrx5 Grx domain. Residues involved in the Prx-Grx contact
of in the hyPrx5 tetramer are labeled and surrounded by a yellow
line. c, the surface of human Prx5. Residues of human Prx5
corresponding to those participating in the Prx-Grx contact of
hyPrx5 are labeled and surrounded by a yellow line. The point of
view in the figure is the same as in a. The orientation was
determined by superposing the two structures (the hyPrx5 Prx
domain and human Prx5). Thr-48, Ser-51, Phe-150, Asp-154, and
Asp-156 of the hyPrx5 Prx domain correspond to Gly-46, Lys-49,
Leu-149, Leu-153, and Pro-155 of human Prx5, respectively (for
the sequence alignment, see Fig. 2). d, the surface of human
Trx. Residues of human Trx involved in the Trx-Trx reductase
contact (PDB code 1F6M) are labeled and surrounded by a yellow
line. The point of view in the figure is the same as in b. The
orientation was determined by superposing the two structures
(the hyPrx5 Grx domain and human Trx). The C  trace of
the NF trace of
the NF  B peptide
bound to human Trx (Ref. 38, PDB code 1MDI) was drawn as a cyan
tube. B peptide
bound to human Trx (Ref. 38, PDB code 1MDI) was drawn as a cyan
tube.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the ASBMB:
J Biol Chem
(2003,
278,
10790-10798)
copyright 2003.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
K.J.Nelson,
S.T.Knutson,
L.Soito,
C.Klomsiri,
L.B.Poole,
and
J.S.Fetrow
(2011).
Analysis of the peroxiredoxin family: Using active-site structure and sequence information for global classification and residue analysis.
|
| |
Proteins,
79,
947-964.
|
 |
|
|
|
|
 |
E.Pedone,
D.Limauro,
K.D'Ambrosio,
G.De Simone,
and
S.Bartolucci
(2010).
Multiple catalytically active thioredoxin folds: a winning strategy for many functions.
|
| |
Cell Mol Life Sci,
67,
3797-3814.
|
 |
|
|
|
|
 |
H.J.Atkinson,
and
P.C.Babbitt
(2009).
An atlas of the thioredoxin fold class reveals the complexity of function-enabling adaptations.
|
| |
PLoS Comput Biol,
5,
e1000541.
|
 |
|
|
|
|
 |
A.Smeets,
E.Loumaye,
A.Clippe,
J.F.Rees,
B.Knoops,
and
J.P.Declercq
(2008).
The crystal structure of the C45S mutant of annelid Arenicola marina peroxiredoxin 6 supports its assignment to the mechanistically typical 2-Cys subfamily without any formation of toroid-shaped decamers.
|
| |
Protein Sci,
17,
700-710.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
F.Gama,
C.Bréhélin,
E.Gelhaye,
Y.Meyer,
J.P.Jacquot,
P.Rey,
and
N.Rouhier
(2008).
Functional analysis and expression characteristics of chloroplastic Prx IIE.
|
| |
Physiol Plant,
133,
599-610.
|
 |
|
|
|
|
 |
K.O.Håkansson,
and
J.R.Winther
(2007).
Structure of glutaredoxin Grx1p C30S mutant from yeast.
|
| |
Acta Crystallogr D Biol Crystallogr,
63,
288-294.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
K.O.Håkansson,
H.Østergaard,
and
J.R.Winther
(2006).
Crystallization of mutant forms of glutaredoxin Grx1p from yeast.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
62,
920-922.
|
 |
|
|
|
|
 |
V.Noguera-Mazon,
I.Krimm,
O.Walker,
and
J.M.Lancelin
(2006).
Protein-protein interactions within peroxiredoxin systems.
|
| |
Photosynth Res,
89,
277-290.
|
 |
|
|
|
|
 |
A.Smeets,
C.Evrard,
M.Landtmeters,
C.Marchand,
B.Knoops,
and
J.P.Declercq
(2005).
Crystal structures of oxidized and reduced forms of human mitochondrial thioredoxin 2.
|
| |
Protein Sci,
14,
2610-2621.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
B.Dombrecht,
C.Heusdens,
S.Beullens,
C.Verreth,
E.Mulkers,
P.Proost,
J.Vanderleyden,
and
J.Michiels
(2005).
Defence of Rhizobium etli bacteroids against oxidative stress involves a complexly regulated atypical 2-Cys peroxiredoxin.
|
| |
Mol Microbiol,
55,
1207-1221.
|
 |
|
|
|
|
 |
S.G.Rhee,
H.Z.Chae,
and
K.Kim
(2005).
Peroxiredoxins: a historical overview and speculative preview of novel mechanisms and emerging concepts in cell signaling.
|
| |
Free Radic Biol Med,
38,
1543-1552.
|
 |
|
|
|
|
 |
T.F.Murphy,
C.Kirkham,
S.Sethi,
and
A.J.Lesse
(2005).
Expression of a peroxiredoxin-glutaredoxin by Haemophilus influenzae in biofilms and during human respiratory tract infection.
|
| |
FEMS Immunol Med Microbiol,
44,
81-89.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

