 |
PDBsum entry 1jtk

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.3.5.4.5
- cytidine deaminase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
|
1.
|
cytidine + H2O + H+ = uridine + NH4+
|
|
2.
|
2'-deoxycytidine + H2O + H+ = 2'-deoxyuridine + NH4+
|
|
 |
 |
 |
 |
 |
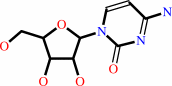 cytidine
cytidine
|
+
|
H2O
|
+
|
H(+)
|
=
|
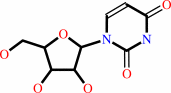 uridine
uridine
|
+
|
NH4(+)
Bound ligand (Het Group name = )
matches with 94.12% similarity
|
|
 |
 |
 |
 |
 |
 2'-deoxycytidine
2'-deoxycytidine
|
+
|
H2O
|
+
|
H(+)
|
=
|
2'-deoxyuridine
Bound ligand (Het Group name = )
corresponds exactly
|
+
|
NH4(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Zn(2+)
|
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Biochemistry
41:2563-2570
(2002)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structure of the tetrameric cytidine deaminase from Bacillus subtilis at 2.0 A resolution.
|
|
E.Johansson,
N.Mejlhede,
J.Neuhard,
S.Larsen.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Cytidine deaminases (CDA, EC 3.5.4.5) are zinc-containing enzymes in the
pyrimidine salvage pathway that catalyze the formation of uridine and
deoxyuridine from cytidine and deoxycytidine, respectively. Two different
classes have been identified in the CDA family, a homodimeric form (D-CDA) with
two zinc ions per dimer and a homotetrameric form (T-CDA) with four zinc ions
per tetramer. We have determined the first structure of a T-CDA from Bacillus
subtilis. The active form of T-CDA is assembled of four identical subunits with
one active site apiece. The subunit of D-CDA is composed of two domains each
exhibiting the same fold as the T-CDA subunits, but only one of them contains
zinc in the active site. The similarity results in a conserved structural core
in the two CDA forms. An intriguing difference between the two CDA structures is
the zinc coordinating residues found at the N-terminal of two alpha-helices:
three cysteine residues in the tetrameric form and two cysteine residues and one
histidine residue in the dimeric form. The role of the zinc ion is to activate a
water molecule and thereby generate a hydroxide ion. How the zinc ion in T-CDA
surrounded with three negatively charged residues can create a similar activity
of T-CDA compared to D-CDA has been an enigma. However, the structure of T-CDA
reveals that the negative charge caused by the three ligands is partly
neutralized by (1) an arginine residue hydrogen-bonded to two of the cysteine
residues and (2) the dipoles of two alpha-helices.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
Z.A.Sánchez-Quitian,
L.F.Timmers,
R.A.Caceres,
J.G.Rehm,
C.E.Thompson,
L.A.Basso,
W.F.de Azevedo,
and
D.S.Santos
(2011).
Crystal structure determination and dynamic studies of Mycobacterium tuberculosis Cytidine deaminase in complex with products.
|
| |
Arch Biochem Biophys,
509,
108-115.
|
 |
|
|
|
|
 |
S.M.Shandilya,
M.N.Nalam,
E.A.Nalivaika,
P.J.Gross,
J.C.Valesano,
K.Shindo,
M.Li,
M.Munson,
W.E.Royer,
E.Harjes,
T.Kono,
H.Matsuo,
R.S.Harris,
M.Somasundaran,
and
C.A.Schiffer
(2010).
Crystal structure of the APOBEC3G catalytic domain reveals potential oligomerization interfaces.
|
| |
Structure,
18,
28-38.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
H.Huthoff,
F.Autore,
S.Gallois-Montbrun,
F.Fraternali,
and
M.H.Malim
(2009).
RNA-dependent oligomerization of APOBEC3G is required for restriction of HIV-1.
|
| |
PLoS Pathog,
5,
e1000330.
|
 |
|
|
|
|
 |
J.Norton,
H.Matsuo,
and
S.J.Sturla
(2009).
Synthesis of deoxytetrahydrouridine.
|
| |
J Org Chem,
74,
2221-2223.
|
 |
|
|
|
|
 |
J.W.Rausch,
L.Chelico,
M.F.Goodman,
and
S.F.Le Grice
(2009).
Dissecting APOBEC3G substrate specificity by nucleoside analog interference.
|
| |
J Biol Chem,
284,
7047-7058.
|
 |
|
|
|
|
 |
M.Henry,
D.Guétard,
R.Suspène,
C.Rusniok,
S.Wain-Hobson,
and
J.P.Vartanian
(2009).
Genetic editing of HBV DNA by monodomain human APOBEC3 cytidine deaminases and the recombinant nature of APOBEC3G.
|
| |
PLoS ONE,
4,
e4277.
|
 |
|
|
|
|
 |
V.E.Marquez,
G.K.Schroeder,
O.R.Ludek,
M.A.Siddiqui,
A.Ezzitouni,
and
R.Wolfenden
(2009).
Contrasting behavior of conformationally locked carbocyclic nucleosides of adenosine and cytidine as substrates for deaminases.
|
| |
Nucleosides Nucleotides Nucleic Acids,
28,
614-632.
|
 |
|
|
|
|
 |
D.Wolf,
and
S.P.Goff
(2008).
Host restriction factors blocking retroviral replication.
|
| |
Annu Rev Genet,
42,
143-163.
|
 |
|
|
|
|
 |
K.M.Chen,
E.Harjes,
P.J.Gross,
A.Fahmy,
Y.Lu,
K.Shindo,
R.S.Harris,
and
H.Matsuo
(2008).
Structure of the DNA deaminase domain of the HIV-1 restriction factor APOBEC3G.
|
| |
Nature,
452,
116-119.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
S.Vincenzetti,
B.Quadrini,
P.Mariani,
G.De Sanctis,
N.Cammertoni,
V.Polzonetti,
S.Pucciarelli,
P.Natalini,
and
A.Vita
(2008).
Modulation of human cytidine deaminase by specific aminoacids involved in the intersubunit interactions.
|
| |
Proteins,
70,
144-156.
|
 |
|
|
|
|
 |
C.Prochnow,
R.Bransteitter,
M.G.Klein,
M.F.Goodman,
and
X.S.Chen
(2007).
The APOBEC-2 crystal structure and functional implications for the deaminase AID.
|
| |
Nature,
445,
447-451.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
T.Kumasaka,
M.Yamamoto,
M.Furuichi,
M.Nakasako,
A.H.Teh,
M.Kimura,
I.Yamaguchi,
and
T.Ueki
(2007).
Crystal Structures of Blasticidin S Deaminase (BSD): IMPLICATIONS FOR DYNAMIC PROPERTIES OF CATALYTIC ZINC.
|
| |
J Biol Chem,
282,
37103-37111.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
H.C.Losey,
A.J.Ruthenburg,
and
G.L.Verdine
(2006).
Crystal structure of Staphylococcus aureus tRNA adenosine deaminase TadA in complex with RNA.
|
| |
Nat Struct Mol Biol,
13,
153-159.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
J.E.Wedekind,
R.Gillilan,
A.Janda,
J.Krucinska,
J.D.Salter,
R.P.Bennett,
J.Raina,
and
H.C.Smith
(2006).
Nanostructures of APOBEC3G support a hierarchical assembly model of high molecular mass ribonucleoprotein particles from dimeric subunits.
|
| |
J Biol Chem,
281,
38122-38126.
|
 |
|
|
|
|
 |
S.C.Chen,
Y.C.Chang,
C.H.Lin,
C.H.Lin,
and
S.H.Liaw
(2006).
Crystal structure of a bifunctional deaminase and reductase from Bacillus subtilis involved in riboflavin biosynthesis.
|
| |
J Biol Chem,
281,
7605-7613.
|
 |
|
|
|
|
 |
H.Huthoff,
and
M.H.Malim
(2005).
Cytidine deamination and resistance to retroviral infection: towards a structural understanding of the APOBEC proteins.
|
| |
Virology,
334,
147-153.
|
 |
|
|
|
|
 |
M.Kuratani,
R.Ishii,
Y.Bessho,
R.Fukunaga,
T.Sengoku,
M.Shirouzu,
S.Sekine,
and
S.Yokoyama
(2005).
Crystal structure of tRNA adenosine deaminase (TadA) from Aquifex aeolicus.
|
| |
J Biol Chem,
280,
16002-16008.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
Y.Qi,
and
N.V.Grishin
(2005).
Structural classification of thioredoxin-like fold proteins.
|
| |
Proteins,
58,
376-388.
|
 |
|
|
|
|
 |
K.Xie,
M.P.Sowden,
G.S.Dance,
A.T.Torelli,
H.C.Smith,
and
J.E.Wedekind
(2004).
The structure of a yeast RNA-editing deaminase provides insight into the fold and function of activation-induced deaminase and APOBEC-1.
|
| |
Proc Natl Acad Sci U S A,
101,
8114-8119.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
R.S.Harris,
and
M.T.Liddament
(2004).
Retroviral restriction by APOBEC proteins.
|
| |
Nat Rev Immunol,
4,
868-877.
|
 |
|
|
|
|
 |
S.H.Liaw,
Y.J.Chang,
C.T.Lai,
H.C.Chang,
and
G.G.Chang
(2004).
Crystal structure of Bacillus subtilis guanine deaminase: the first domain-swapped structure in the cytidine deaminase superfamily.
|
| |
J Biol Chem,
279,
35479-35485.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
X.I.Ambroggio,
D.C.Rees,
and
R.J.Deshaies
(2004).
JAMM: a metalloprotease-like zinc site in the proteasome and signalosome.
|
| |
PLoS Biol,
2,
E2.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
Y.J.Chang,
C.H.Huang,
C.Y.Hu,
and
S.H.Liaw
(2004).
Crystallization and preliminary crystallographic analysis of Bacillus subtilis guanine deaminase.
|
| |
Acta Crystallogr D Biol Crystallogr,
60,
1152-1154.
|
 |
|
|
|
|
 |
E.Johansson,
O.Bjornberg,
P.O.Nyman,
and
S.Larsen
(2003).
Structure of the bifunctional dCTP deaminase-dUTPase from Methanocaldococcus jannaschii and its relation to other homotrimeric dUTPases.
|
| |
J Biol Chem,
278,
27916-27922.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
G.C.Ireton,
M.E.Black,
and
B.L.Stoddard
(2003).
The 1.14 A crystal structure of yeast cytosine deaminase: evolution of nucleotide salvage enzymes and implications for genetic chemotherapy.
|
| |
Structure,
11,
961-972.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
H.J.Tran,
M.D.Allen,
J.Löwe,
and
M.Bycroft
(2003).
Structure of the Jab1/MPN domain and its implications for proteasome function.
|
| |
Biochemistry,
42,
11460-11465.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
H.Li,
H.Xu,
D.E.Graham,
and
R.H.White
(2003).
The Methanococcus jannaschii dCTP deaminase is a bifunctional deaminase and diphosphatase.
|
| |
J Biol Chem,
278,
11100-11106.
|
 |
|
|
|
|
 |
J.E.Wedekind,
G.S.Dance,
M.P.Sowden,
and
H.C.Smith
(2003).
Messenger RNA editing in mammals: new members of the APOBEC family seeking roles in the family business.
|
| |
Trends Genet,
19,
207-216.
|
 |
|
|
|
|
 |
T.P.Ko,
J.J.Lin,
C.Y.Hu,
Y.H.Hsu,
A.H.Wang,
and
S.H.Liaw
(2003).
Crystal structure of yeast cytosine deaminase. Insights into enzyme mechanism and evolution.
|
| |
J Biol Chem,
278,
19111-19117.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
T.X.Xiang,
R.Niemi,
P.Bummer,
and
B.D.Anderson
(2003).
Epimer interconversion, isomerization, and hydrolysis of tetrahydrouridine: implications for cytidine deaminase inhibition.
|
| |
J Pharm Sci,
92,
2027-2039.
|
 |
|
|
|
|
 |
Y.H.Hsu,
C.Y.Hu,
J.J.Lin,
and
S.H.Liaw
(2003).
Crystallization and preliminary crystallographic analysis of yeast cytosine deaminase.
|
| |
Acta Crystallogr D Biol Crystallogr,
59,
950-952.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

