 |
PDBsum entry 1hgx

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Transferase (glycosyltransferase)
|
PDB id
|
|

|

|
1hgx
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.2.4.2.22
- xanthine phosphoribosyltransferase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
XMP + diphosphate = xanthine + 5-phospho-alpha-D-ribose 1-diphosphate
|
 |
 |
 |
 |
 |
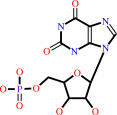 XMP
XMP
|
+
|
diphosphate
Bound ligand (Het Group name = )
matches with 92.00% similarity
|
=
|
 xanthine
xanthine
|
+
|
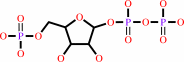 5-phospho-alpha-D-ribose 1-diphosphate
5-phospho-alpha-D-ribose 1-diphosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 3:
|
 |
E.C.2.4.2.8
- hypoxanthine phosphoribosyltransferase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
IMP + diphosphate = hypoxanthine + 5-phospho-alpha-D-ribose 1-diphosphate
|
 |
 |
 |
 |
 |
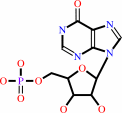 IMP
IMP
|
+
|
diphosphate
Bound ligand (Het Group name = )
matches with 95.83% similarity
|
=
|
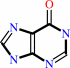 hypoxanthine
hypoxanthine
|
+
|
 5-phospho-alpha-D-ribose 1-diphosphate
5-phospho-alpha-D-ribose 1-diphosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Biochemistry
35:7032-7040
(1996)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structure of the hypoxanthine-guanine-xanthine phosphoribosyltransferase from the protozoan parasite Tritrichomonas foetus.
|
|
J.R.Somoza,
M.S.Chin,
P.J.Focia,
C.C.Wang,
R.J.Fletterick.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The crystal structure of the hypoxanthine-guanine-xanthine
phosphoribosyltransferase (HGXPRTase) from Tritrichomonas foetus has been
determined and refined against X-ray data to 1.9 A resolution. T. foetus
HGXPRTase crystallizes as an asymmetric dimer, with GMP bound to only one of the
two molecules that form the asymmetric unit. Each molecule of HGXPRTase is
formed by two lobes joined by a short "hinge" region, and the GMP binds in a
cavity between the two lobes. A comparison of the two molecules in the
asymmetric unit shows that the hinge region is flexible and that ligand binding
affects the relative positions of the two lobes. The binding of GMP brings the
two lobes closer together, rotating one lobe by about 5 degrees relative to the
other. T. foetus appears to depend on HGXPRTase for its supply of GMP, making
this enzyme a target for antiparasite drug design. A comparison of the
structures of T. foetus HGXPRTase and human HGPRTase reveals that, while these
enzymes retain a similar polypeptide fold, there are substantial differences
between the active sites of these two homologs. These differences suggest that
it will be possible to find compounds that selectively inhibit the parasite
enzyme.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
H.Deng,
R.Callender,
V.L.Schramm,
and
C.Grubmeyer
(2010).
Pyrophosphate activation in hypoxanthine--guanine phosphoribosyltransferase with transition state analogue.
|
| |
Biochemistry,
49,
2705-2714.
|
 |
|
|
|
|
 |
P.Gayathri,
I.N.Sujay Subbayya,
C.S.Ashok,
T.S.Selvi,
H.Balaram,
and
M.R.Murthy
(2008).
Crystal structure of a chimera of human and Plasmodium falciparum hypoxanthine guanine phosphoribosyltransferases provides insights into oligomerization.
|
| |
Proteins,
73,
1010-1020.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
P.S.Monzani,
S.Trapani,
O.H.Thiemann,
and
G.Oliva
(2007).
Crystal structure of Leishmania tarentolae hypoxanthine-guanine phosphoribosyltransferase.
|
| |
BMC Struct Biol,
7,
59.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
Q.Chen,
D.You,
Y.Liang,
X.Su,
X.Gu,
M.Luo,
and
X.Zheng
(2007).
Crystal structure of Thermoanaerobacter tengcongensis hypoxanthine-guanine phosphoribosyl transferase L160I mutant--insights into inhibitor design.
|
| |
FEBS J,
274,
4408-4415.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
X.Liu,
W.Qian,
X.Liu,
H.Qin,
and
D.Wang
(2007).
Molecular and functional analysis of hypoxanthine-guanine phosphoribosyltransferase from Arabidopsis thaliana.
|
| |
New Phytol,
175,
448-461.
|
 |
|
|
|
|
 |
M.Duckworth,
A.Ménard,
F.Megraud,
and
G.L.Mendz
(2006).
Bioinformatic analysis of Helicobacter pylori XGPRTase: a potential therapeutic target.
|
| |
Helicobacter,
11,
287-295.
|
 |
|
|
|
|
 |
J.Duan,
L.Nilsson,
and
B.Lambert
(2004).
Structural and functional analysis of mutations at the human hypoxanthine phosphoribosyl transferase (HPRT1) locus.
|
| |
Hum Mutat,
23,
599-611.
|
 |
|
|
|
|
 |
R.V.Dumitru,
and
S.W.Ragsdale
(2004).
Mechanism of 4-(beta-D-ribofuranosyl)aminobenzene 5'-phosphate synthase, a key enzyme in the methanopterin biosynthetic pathway.
|
| |
J Biol Chem,
279,
39389-39395.
|
 |
|
|
|
|
 |
D.You,
Q.Chen,
Y.Liang,
J.An,
R.Li,
X.Gu,
M.Luo,
and
X.D.Su
(2003).
Protein preparation, crystallization and preliminary X-ray crystallographic studies of a thermostable hypoxanthine-guanine phosphoribosyltransferase from Thermoanaerobacter tengcongensis.
|
| |
Acta Crystallogr D Biol Crystallogr,
59,
1863-1865.
|
 |
|
|
|
|
 |
A.E.Sarver,
and
C.C.Wang
(2002).
The adenine phosphoribosyltransferase from Giardia lamblia has a unique reaction mechanism and unusual substrate binding properties.
|
| |
J Biol Chem,
277,
39973-39980.
|
 |
|
|
|
|
 |
A.Kadziola,
J.Neuhard,
and
S.Larsen
(2002).
Structure of product-bound Bacillus caldolyticus uracil phosphoribosyltransferase confirms ordered sequential substrate binding.
|
| |
Acta Crystallogr D Biol Crystallogr,
58,
936-945.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
L.J.Wallace,
D.Candlish,
and
H.P.De Koning
(2002).
Different substrate recognition motifs of human and trypanosome nucleobase transporters. Selective uptake of purine antimetabolites.
|
| |
J Biol Chem,
277,
26149-26156.
|
 |
|
|
|
|
 |
M.A.Schumacher,
C.J.Bashor,
M.H.Song,
K.Otsu,
S.Zhu,
R.J.Parry,
B.Ullman,
and
R.G.Brennan
(2002).
The structural mechanism of GTP stabilized oligomerization and catalytic activation of the Toxoplasma gondii uracil phosphoribosyltransferase.
|
| |
Proc Natl Acad Sci U S A,
99,
78-83.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
A.M.Aronov,
N.R.Munagala,
I.D.Kuntz,
and
C.C.Wang
(2001).
Virtual screening of combinatorial libraries across a gene family: in search of inhibitors of Giardia lamblia guanine phosphoribosyltransferase.
|
| |
Antimicrob Agents Chemother,
45,
2571-2576.
|
 |
|
|
|
|
 |
B.Canyuk,
P.J.Focia,
and
A.E.Eakin
(2001).
The role for an invariant aspartic acid in hypoxanthine phosphoribosyltransferases is examined using saturation mutagenesis, functional analysis, and X-ray crystallography.
|
| |
Biochemistry,
40,
2754-2765.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
E.R.Bonner,
J.N.D'Elia,
B.K.Billips,
and
R.L.Switzer
(2001).
Molecular recognition of pyr mRNA by the Bacillus subtilis attenuation regulatory protein PyrR.
|
| |
Nucleic Acids Res,
29,
4851-4865.
|
 |
|
|
|
|
 |
N.Munagala,
V.J.Basus,
and
C.C.Wang
(2001).
Role of the flexible loop of hypoxanthine-guanine-xanthine phosphoribosyltransferase from Tritrichomonas foetus in enzyme catalysis.
|
| |
Biochemistry,
40,
4303-4311.
|
 |
|
|
|
|
 |
A.M.Aronov,
N.R.Munagala,
P.R.Ortiz De Montellano,
I.D.Kuntz,
and
C.C.Wang
(2000).
Rational design of selective submicromolar inhibitors of Tritrichomonas foetus hypoxanthine-guanine-xanthine phosphoribosyltransferase.
|
| |
Biochemistry,
39,
4684-4691.
|
 |
|
|
|
|
 |
A.Héroux,
E.L.White,
L.J.Ross,
and
D.W.Borhani
(1999).
Crystal structures of the Toxoplasma gondii hypoxanthine-guanine phosphoribosyltransferase-GMP and -IMP complexes: comparison of purine binding interactions with the XMP complex.
|
| |
Biochemistry,
38,
14485-14494.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
A.Héroux,
E.L.White,
L.J.Ross,
R.L.Davis,
and
D.W.Borhani
(1999).
Crystal structure of Toxoplasma gondii hypoxanthine-guanine phosphoribosyltransferase with XMP, pyrophosphate, and two Mg(2+) ions bound: insights into the catalytic mechanism.
|
| |
Biochemistry,
38,
14495-14506.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
A.Jardim,
S.E.Bergeson,
S.Shih,
N.Carter,
R.W.Lucas,
G.Merlin,
P.J.Myler,
K.Stuart,
and
B.Ullman
(1999).
Xanthine phosphoribosyltransferase from Leishmania donovani. Molecular cloning, biochemical characterization, and genetic analysis.
|
| |
J Biol Chem,
274,
34403-34410.
|
 |
|
|
|
|
 |
C.L.Phillips,
B.Ullman,
R.G.Brennan,
and
C.P.Hill
(1999).
Crystal structures of adenine phosphoribosyltransferase from Leishmania donovani.
|
| |
EMBO J,
18,
3533-3545.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
E.M.Khalil,
J.De Angelis,
M.Ishii,
and
P.A.Cole
(1999).
Mechanism-based inhibition of the melatonin rhythm enzyme: pharmacologic exploitation of active site functional plasticity.
|
| |
Proc Natl Acad Sci U S A,
96,
12418-12423.
|
 |
|
|
|
|
 |
G.K.Balendiran,
J.A.Molina,
Y.Xu,
J.Torres-Martinez,
R.Stevens,
P.J.Focia,
A.E.Eakin,
J.C.Sacchettini,
and
S.P.Craig
(1999).
Ternary complex structure of human HGPRTase, PRPP, Mg2+, and the inhibitor HPP reveals the involvement of the flexible loop in substrate binding.
|
| |
Protein Sci,
8,
1023-1031.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
J.P.Page,
N.R.Munagala,
and
C.C.Wang
(1999).
Point mutations in the guanine phosphoribosyltransferase from Giardia lamblia modulate pyrophosphate binding and enzyme catalysis.
|
| |
Eur J Biochem,
259,
565-571.
|
 |
|
|
|
|
 |
J.Sauer,
and
P.Nygaard
(1999).
Expression of the Methanobacterium thermoautotrophicum hpt gene, encoding hypoxanthine (Guanine) phosphoribosyltransferase, in Escherichia coli.
|
| |
J Bacteriol,
181,
1958-1962.
|
 |
|
|
|
|
 |
C.C.Lee,
S.P.Craig,
and
A.E.Eakin
(1998).
A single amino acid substitution in the human and a bacterial hypoxanthine phosphoribosyltransferase modulates specificity for the binding of guanine.
|
| |
Biochemistry,
37,
3491-3498.
|
 |
|
|
|
|
 |
C.R.Muchmore,
J.M.Krahn,
J.H.Kim,
H.Zalkin,
and
J.L.Smith
(1998).
Crystal structure of glutamine phosphoribosylpyrophosphate amidotransferase from Escherichia coli.
|
| |
Protein Sci,
7,
39-51.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
J.L.Smith
(1998).
Glutamine PRPP amidotransferase: snapshots of an enzyme in action.
|
| |
Curr Opin Struct Biol,
8,
686-694.
|
 |
|
|
|
|
 |
J.R.Somoza,
A.G.Skillman,
N.R.Munagala,
C.M.Oshiro,
R.M.Knegtel,
S.Mpoke,
R.J.Fletterick,
I.D.Kuntz,
and
C.C.Wang
(1998).
Rational design of novel antimicrobials: blocking purine salvage in a parasitic protozoan.
|
| |
Biochemistry,
37,
5344-5348.
|
 |
|
|
|
|
 |
M.A.Schumacher,
D.Carter,
D.M.Scott,
D.S.Roos,
B.Ullman,
and
R.G.Brennan
(1998).
Crystal structures of Toxoplasma gondii uracil phosphoribosyltransferase reveal the atomic basis of pyrimidine discrimination and prodrug binding.
|
| |
EMBO J,
17,
3219-3232.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
N.R.Munagala,
and
C.C.Wang
(1998).
Altering the purine specificity of hypoxanthine-guanine-xanthine phosphoribosyltransferase from Tritrichomonas foetus by structure-based point mutations in the enzyme protein.
|
| |
Biochemistry,
37,
16612-16619.
|
 |
|
|
|
|
 |
N.R.Munagala,
M.S.Chin,
and
C.C.Wang
(1998).
Steady-state kinetics of the hypoxanthine-guanine-xanthine phosphoribosyltransferase from Tritrichomonas foetus: the role of threonine-47.
|
| |
Biochemistry,
37,
4045-4051.
|
 |
|
|
|
|
 |
P.J.Focia,
S.P.Craig,
and
A.E.Eakin
(1998).
Approaching the transition state in the crystal structure of a phosphoribosyltransferase.
|
| |
Biochemistry,
37,
17120-17127.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
P.J.Focia,
S.P.Craig,
R.Nieves-Alicea,
R.J.Fletterick,
and
A.E.Eakin
(1998).
A 1.4 A crystal structure for the hypoxanthine phosphoribosyltransferase of Trypanosoma cruzi.
|
| |
Biochemistry,
37,
15066-15075.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
R.Nieves-Alicea,
P.J.Focia,
S.P.Craig,
and
A.E.Eakin
(1998).
Limited proteolysis of a trypanosomal hypoxanthine phosphoribosyltransferase yields crystals that diffract X-rays to near atomic resolution.
|
| |
Biochim Biophys Acta,
1388,
500-505.
|
 |
|
|
|
|
 |
A.E.Eakin,
A.Guerra,
P.J.Focia,
J.Torres-Martinez,
and
S.P.Craig
(1997).
Hypoxanthine phosphoribosyltransferase from Trypanosoma cruzi as a target for structure-based inhibitor design: crystallization and inhibition studies with purine analogs.
|
| |
Antimicrob Agents Chemother,
41,
1686-1692.
|
 |
|
|
|
|
 |
F.G.Whitby,
H.Luecke,
P.Kuhn,
J.R.Somoza,
J.A.Huete-Perez,
J.D.Phillips,
C.P.Hill,
R.J.Fletterick,
and
C.C.Wang
(1997).
Crystal structure of Tritrichomonas foetus inosine-5'-monophosphate dehydrogenase and the enzyme-product complex.
|
| |
Biochemistry,
36,
10666-10674.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
J.Kanaani,
D.Maltby,
J.R.Somoza,
and
C.C.Wang
(1997).
Inactivation of Tritrichomonas foetus and Schistosoma mansoni purine phosphoribosyltransferases by arginine-specific reagents.
|
| |
Eur J Biochem,
244,
810-817.
|
 |
|
|
|
|
 |
J.M.Krahn,
J.H.Kim,
M.R.Burns,
R.J.Parry,
H.Zalkin,
and
J.L.Smith
(1997).
Coupled formation of an amidotransferase interdomain ammonia channel and a phosphoribosyltransferase active site.
|
| |
Biochemistry,
36,
11061-11068.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
S.Chen,
D.R.Tomchick,
D.Wolle,
P.Hu,
J.L.Smith,
R.L.Switzer,
and
H.Zalkin
(1997).
Mechanism of the synergistic end-product regulation of Bacillus subtilis glutamine phosphoribosylpyrophosphate amidotransferase by nucleotides.
|
| |
Biochemistry,
36,
10718-10726.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
S.Vos,
J.de Jersey,
and
J.L.Martin
(1997).
Crystal structure of Escherichia coli xanthine phosphoribosyltransferase.
|
| |
Biochemistry,
36,
4125-4134.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
Y.Xu,
J.Eads,
J.C.Sacchettini,
and
C.Grubmeyer
(1997).
Kinetic mechanism of human hypoxanthine-guanine phosphoribosyltransferase: rapid phosphoribosyl transfer chemistry.
|
| |
Biochemistry,
36,
3700-3712.
|
 |
|
|
|
|
 |
J.Kanaani,
J.R.Somoza,
D.Maltby,
and
C.C.Wang
(1996).
Probing the active site of Tritrichomonas foetus hypoxanthine-guanine-xanthine phosphoribosyltransferase using covalent modification of cysteine residues.
|
| |
Eur J Biochem,
239,
764-772.
|
 |
|
|
|
|
 |
M.A.Schumacher,
D.Carter,
D.S.Ross,
B.Ullman,
and
R.G.Brennan
(1996).
Crystal structures of Toxoplasma gondii HGXPRTase reveal the catalytic role of a long flexible loop.
|
| |
Nat Struct Biol,
3,
881-887.
|
 |
|
PDB code:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

