 |
PDBsum entry 1h7w

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Electron transfer
|
PDB id
|
|

|

|
1h7w
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Electron transfer
|
 |
|
Title:
|
 |
Dihydropyrimidine dehydrogenase (dpd) from pig
|
|
Structure:
|
 |
Dihydropyrimidine dehydrogenase. Chain: a, b, c, d. Synonym: dihydrouracil dehydrogenase, dihydrothymine dehydrogenase. Engineered: yes
|
|
Source:
|
 |
Sus scrofa. Wild boar. Organism_taxid: 9823. Cellular_location: cytoplasm. Gene: dpyd. Expressed in: escherichia coli dh5[alpha]. Expression_system_taxid: 668369.
|
|
Biol. unit:
|
 |
Homo-Dimer (from PDB file)
|
|
Resolution:
|
 |
|
1.90Å
|
R-factor:
|
0.174
|
R-free:
|
0.196
|
|
|
Authors:
|
 |
D.Dobritzsch,G.Schneider,K.D.Schnackerz,Y.Lindqvist
|
Key ref:

|
 |
D.Dobritzsch
et al.
(2001).
Crystal structure of dihydropyrimidine dehydrogenase, a major determinant of the pharmacokinetics of the anti-cancer drug 5-fluorouracil.
EMBO J,
20,
650-660.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
19-Jan-01
|
Release date:
|
23-Feb-01
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
Q28943
(DPYD_PIG) -
Dihydropyrimidine dehydrogenase [NADP(+)] from Sus scrofa
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
1025 a.a.
1016 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|
*
PDB and UniProt seqs differ
at 1 residue position (black
cross)
|
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.3.1.2
- dihydropyrimidine dehydrogenase (NADP(+)).
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
5,6-dihydrouracil + NADP+ = uracil + NADPH + H+
|
 |
 |
 |
 |
 |
 5,6-dihydrouracil
5,6-dihydrouracil
|
+
|
NADP(+)
Bound ligand (Het Group name = )
matches with 71.19% similarity
|
=
|
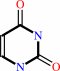 uracil
uracil
|
+
|
 NADPH
NADPH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
EMBO J
20:650-660
(2001)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structure of dihydropyrimidine dehydrogenase, a major determinant of the pharmacokinetics of the anti-cancer drug 5-fluorouracil.
|
|
D.Dobritzsch,
G.Schneider,
K.D.Schnackerz,
Y.Lindqvist.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Dihydropyrimidine dehydrogenase catalyzes the first step in pyrimidine
degradation: the NADPH-dependent reduction of uracil and thymine to the
corresponding 5,6-dihydropyrimidines. Its controlled inhibition has become an
adjunct target for cancer therapy, since the enzyme is also responsible for the
rapid breakdown of the chemotherapeutic drug 5-fluorouracil. The crystal
structure of the homodimeric pig liver enzyme (2x 111 kDa) determined at 1.9 A
resolution reveals a highly modular subunit organization, consisting of five
domains with different folds. Dihydropyrimidine dehydrogenase contains two FAD,
clusters, arranged in two electron transfer chains
that pass the dimer interface twice. Two of the Fe-S clusters show a hitherto
unobserved coordination involving a glutamine residue. The ternary complex of an
inactive mutant of the enzyme with bound NADPH and 5-fluorouracil reveals the
architecture of the substrate-binding sites and residues responsible for
recognition and binding of the drug.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
Figure 1 Structure of pig liver DPD. (A) Schematic view of the
subunit of DPD with the domains in different colors. The
cofactors are shown as ball-and-stick models, iron ions in
magenta and sulfur atoms in green. (B) The DPD dimer. The color
codes for the domains of the first subunit are the same as in
(A), the corresponding domains in the second subunit are shown
in light green, brown, cyan, pink and light blue.
|
 |
Figure 6.
Figure 6 Electron transfer pathways in DPD. Distances between
closest atoms of the cofactors are indicated. The nicotinamide
ring of NADPH is shown at its assumed position during electron
transfer.
|
 |
|
|

|
| |
The above figures are
reprinted
from an Open Access publication published by Macmillan Publishers Ltd:
EMBO J
(2001,
20,
650-660)
copyright 2001.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
A.B.van Kuilenburg,
J.Meijer,
A.N.Mul,
R.Meinsma,
V.Schmid,
D.Dobritzsch,
R.C.Hennekam,
M.M.Mannens,
M.Kiechle,
M.C.Etienne-Grimaldi,
H.J.Klümpen,
J.G.Maring,
V.A.Derleyn,
E.Maartense,
G.Milano,
R.Vijzelaar,
and
E.Gross
(2010).
Intragenic deletions and a deep intronic mutation affecting pre-mRNA splicing in the dihydropyrimidine dehydrogenase gene as novel mechanisms causing 5-fluorouracil toxicity.
|
| |
Hum Genet,
128,
529-538.
|
 |
|
|
|
|
 |
A.Karcher,
A.Schele,
and
K.P.Hopfner
(2008).
X-ray structure of the complete ABC enzyme ABCE1 from Pyrococcus abyssi.
|
| |
J Biol Chem,
283,
7962-7971.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
E.Gross,
B.Busse,
M.Riemenschneider,
S.Neubauer,
K.Seck,
H.G.Klein,
M.Kiechle,
F.Lordick,
and
A.Meindl
(2008).
Strong association of a common dihydropyrimidine dehydrogenase gene polymorphism with fluoropyrimidine-related toxicity in cancer patients.
|
| |
PLoS ONE,
3,
e4003.
|
 |
|
|
|
|
 |
M.A.Vanoni,
and
B.Curti
(2008).
Structure-function studies of glutamate synthases: a class of self-regulated iron-sulfur flavoenzymes essential for nitrogen assimilation.
|
| |
IUBMB Life,
60,
287-300.
|
 |
|
|
|
|
 |
M.Cottevieille,
E.Larquet,
S.Jonic,
M.V.Petoukhov,
G.Caprini,
S.Paravisi,
D.I.Svergun,
M.A.Vanoni,
and
N.Boisset
(2008).
The subnanometer resolution structure of the glutamate synthase 1.2-MDa hexamer by cryoelectron microscopy and its oligomerization behavior in solution: functional implications.
|
| |
J Biol Chem,
283,
8237-8249.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
S.Jonić,
C.O.Sorzano,
and
N.Boisset
(2008).
Comparison of single-particle analysis and electron tomography approaches: an overview.
|
| |
J Microsc,
232,
562-579.
|
 |
|
|
|
|
 |
C.O.Sorzano,
S.Jonic,
M.Cottevieille,
E.Larquet,
N.Boisset,
and
S.Marco
(2007).
3D electron microscopy of biological nanomachines: principles and applications.
|
| |
Eur Biophys J,
36,
995.
|
 |
|
|
|
|
 |
X.Zhang,
and
R.B.Diasio
(2007).
Regulation of human dihydropyrimidine dehydrogenase: implications in the pharmacogenetics of 5-FU-based chemotherapy.
|
| |
Pharmacogenomics,
8,
257-265.
|
 |
|
|
|
|
 |
X.Zhang,
R.Soong,
K.Wang,
L.Li,
J.R.Davie,
V.Guarcello,
and
R.B.Diasio
(2007).
Suppression of DPYD expression in RKO cells via DNA methylation in the regulatory region of the DPYD promoter: a potentially important epigenetic mechanism regulating DPYD expression.
|
| |
Biochem Cell Biol,
85,
337-346.
|
 |
|
|
|
|
 |
J.M.Rawls
(2006).
Analysis of pyrimidine catabolism in Drosophila melanogaster using epistatic interactions with mutations of pyrimidine biosynthesis and beta-alanine metabolism.
|
| |
Genetics,
172,
1665-1674.
|
 |
|
|
|
|
 |
S.S.Krishna,
R.I.Sadreyev,
and
N.V.Grishin
(2006).
A tale of two ferredoxins: sequence similarity and structural differences.
|
| |
BMC Struct Biol,
6,
8.
|
 |
|
|
|
|
 |
A.B.Van Kuilenburg,
R.Meinsma,
E.Beke,
B.Bobba,
P.Boffi,
G.M.Enns,
D.R.Witt,
and
D.Dobritzsch
(2005).
Identification of three novel mutations in the dihydropyrimidine dehydrogenase gene associated with altered pre-mRNA splicing or protein function.
|
| |
Biol Chem,
386,
319-324.
|
 |
|
|
|
|
 |
A.Somanchi,
D.Barnes,
and
S.P.Mayfield
(2005).
A nuclear gene of Chlamydomonas reinhardtii, Tba1, encodes a putative oxidoreductase required for translation of the chloroplast psbA mRNA.
|
| |
Plant J,
42,
341-352.
|
 |
|
|
|
|
 |
M.A.Vanoni,
L.Dossena,
R.H.van den Heuvel,
and
B.Curti
(2005).
Structure-function studies on the complex iron-sulfur flavoprotein glutamate synthase: the key enzyme of ammonia assimilation.
|
| |
Photosynth Res,
83,
219-238.
|
 |
|
|
|
|
 |
M.H.Hefti,
J.Vervoort,
and
W.J.van Berkel
(2003).
Deflavination and reconstitution of flavoproteins.
|
| |
Eur J Biochem,
270,
4227-4242.
|
 |
|
|
|
|
 |
M.V.Petoukhov,
D.I.Svergun,
P.V.Konarev,
S.Ravasio,
R.H.van den Heuvel,
B.Curti,
and
M.A.Vanoni
(2003).
Quaternary structure of Azospirillum brasilense NADPH-dependent glutamate synthase in solution as revealed by synchrotron radiation x-ray scattering.
|
| |
J Biol Chem,
278,
29933-29939.
|
 |
|
|
|
|
 |
P.A.Hubbard,
X.Liang,
H.Schulz,
and
J.J.Kim
(2003).
The crystal structure and reaction mechanism of Escherichia coli 2,4-dienoyl-CoA reductase.
|
| |
J Biol Chem,
278,
37553-37560.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
C.A.Bottoms,
P.E.Smith,
and
J.J.Tanner
(2002).
A structurally conserved water molecule in Rossmann dinucleotide-binding domains.
|
| |
Protein Sci,
11,
2125-2137.
|
 |
|
|
|
|
 |
D.Dobritzsch,
S.Ricagno,
G.Schneider,
K.D.Schnackerz,
and
Y.Lindqvist
(2002).
Crystal structure of the productive ternary complex of dihydropyrimidine dehydrogenase with NADPH and 5-iodouracil. Implications for mechanism of inhibition and electron transfer.
|
| |
J Biol Chem,
277,
13155-13166.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
L.K.Mattison,
M.R.Johnson,
and
R.B.Diasio
(2002).
A comparative analysis of translated dihydropyrimidine dehydrogenase cDNA; conservation of functional domains and relevance to genetic polymorphisms.
|
| |
Pharmacogenetics,
12,
133-144.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

