 |
PDBsum entry 1gin

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Ligase
|
 |
|
Title:
|
 |
Crystal structure of adenylosuccinate synthetase from escherichia coli complexed with gdp, imp, hadacidin, no3-, and mg2+. Data collected at 298k (ph 6.5).
|
|
Structure:
|
 |
Adenylosuccinate synthetase. Chain: a. Ec: 6.3.4.4
|
|
Source:
|
 |
Escherichia coli. Organism_taxid: 562. Strain: pur a strain h1238. Atcc: coli genetic stock center, strain number 5408. Other_details: gift from dr. B. Bachman (genetic center, yale university)
|
|
Biol. unit:
|
 |
 Dimer (from PDB file)
Dimer (from PDB file)
|
|
Resolution:
|
 |
|
2.80Å
|
R-factor:
|
0.188
|
R-free:
|
0.232
|
|
|
Authors:
|
 |
B.W.Poland,H.J.Fromm,R.B.Honzatko
|
Key ref:

|
 |
B.W.Poland
et al.
(1996).
Crystal structures of adenylosuccinate synthetase from Escherichia coli complexed with GDP, IMP hadacidin, NO3-, and Mg2+.
J Mol Biol,
264,
1013-1027.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
18-Jun-96
|
Release date:
|
12-Feb-97
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P0A7D4
(PURA_ECOLI) -
Adenylosuccinate synthetase from Escherichia coli (strain K12)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
432 a.a.
431 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.6.3.4.4
- adenylosuccinate synthase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
AMP and GMP Biosynthesis
|
 |
 |
 |
 |
 |
Reaction:
|
 |
IMP + L-aspartate + GTP = N6-(1,2-dicarboxyethyl)-AMP + GDP + phosphate + 2 H+
|
 |
 |
 |
 |
 |
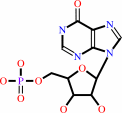 IMP
IMP
|
+
|
L-aspartate
Bound ligand (Het Group name = )
corresponds exactly
|
+
|
GTP
Bound ligand (Het Group name = )
matches with 41.67% similarity
|
=
|
N(6)-(1,2-dicarboxyethyl)-AMP
Bound ligand (Het Group name = )
corresponds exactly
|
+
|
 GDP
GDP
|
+
|
 phosphate
phosphate
|
+
|
2
×
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Mol Biol
264:1013-1027
(1996)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structures of adenylosuccinate synthetase from Escherichia coli complexed with GDP, IMP hadacidin, NO3-, and Mg2+.
|
|
B.W.Poland,
H.J.Fromm,
R.B.Honzatko.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Crystal structures of adenylosuccinate synthetase from Esherichia coli complexed
with Mg2+, IMP, GDP, NO3- and hadacidin at 298 and 100 K have been refined to
R-factors of 0.188 and 0.206 against data to 2.8 A and 2.5 A resolution,
respectively. Conformational changes of up to 9 A relative to the unligated
enzyme occur in loops that bind to Mg2+, GDP, IMP and hadacidin. Mg2+ binds
directly to GDP, NO3-, hadacidin and the protein, but is only five-coordinated.
Asp13, which approaches, but does not occupy the sixth coordination site of
Mg2+, hydrogen bonds to N1 of IMP. The nitrogen atom of NO3- is approximately
2.7 A from O6 of IMP, reflecting a strong electrostatic interaction between the
electron-deficient nitrogen atom and the electron-rich O6. The spatial
relationships between GDP, NO3- and Mg2+ suggest an interaction between the
beta,gamma-bridging oxygen atom of GTP and Mg2+ in the enzyme-substrate complex.
His41 hydrogen bonds to the beta-phosphate group of GDP and approaches bound
NO3-. The aldehyde group of hadacidin coordinates to the Mg2+, while its
carboxyl group interacts with backbone amide groups 299 to 303 and the
side-chain of Arg303. The 5'-phosphate group of IMP interacts with Asn38,
Thr129, Thr239 and Arg143 (from a monomer related by 2-fold symmetry). A
mechanism is proposed for the two-step reaction governed by the synthetase, in
which His41 and Asp13 are essential catalytic side-chains.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
Figure 1. A diagram of hadacidin showing chirality,
covalent linkages and names of atoms (in parentheses).
Groups within circles are represented by single, unified
atoms in refinement.
|
 |
Figure 8.
Figure 8. Stereoview of Mg
2+
in
the active site of the ligated syn-
thetase at 100 K. Broken lines rep-
resent donor-acceptor interactions,
coordinate bonds or significant elec-
trostatic interactions less than 3.2 Å .
Ligands are drawn with bold lines.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from Elsevier:
J Mol Biol
(1996,
264,
1013-1027)
copyright 1996.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
D.L.Scott,
G.Diez,
and
W.H.Goldmann
(2006).
Protein-lipid interactions: correlation of a predictive algorithm for lipid-binding sites with three-dimensional structural data.
|
| |
Theor Biol Med Model,
3,
17.
|
 |
|
|
|
|
 |
A.Yamashita,
K.Maeda,
and
Y.Maéda
(2003).
Crystal structure of CapZ: structural basis for actin filament barbed end capping.
|
| |
EMBO J,
22,
1529-1538.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
T.Borza,
C.V.Iancu,
E.Pike,
R.B.Honzatko,
and
H.J.Fromm
(2003).
Variations in the response of mouse isozymes of adenylosuccinate synthetase to inhibitors of physiological relevance.
|
| |
J Biol Chem,
278,
6673-6679.
|
 |
|
|
|
|
 |
A.Gorrell,
W.Wang,
E.Underbakke,
Z.Hou,
R.B.Honzatko,
and
H.J.Fromm
(2002).
Determinants of L-aspartate and IMP recognition in Escherichia coli adenylosuccinate synthetase.
|
| |
J Biol Chem,
277,
8817-8821.
|
 |
|
|
|
|
 |
C.V.Iancu,
T.Borza,
H.J.Fromm,
and
R.B.Honzatko
(2002).
IMP, GTP, and 6-phosphoryl-IMP complexes of recombinant mouse muscle adenylosuccinate synthetase.
|
| |
J Biol Chem,
277,
26779-26787.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
C.V.Iancu,
T.Borza,
H.J.Fromm,
and
R.B.Honzatko
(2002).
Feedback inhibition and product complexes of recombinant mouse muscle adenylosuccinate synthetase.
|
| |
J Biol Chem,
277,
40536-40543.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
Z.Hou,
W.Wang,
H.J.Fromm,
and
R.B.Honzatko
(2002).
IMP Alone Organizes the Active Site of Adenylosuccinate Synthetase from Escherichia coli.
|
| |
J Biol Chem,
277,
5970-5976.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
C.V.Iancu,
T.Borza,
J.Y.Choe,
H.J.Fromm,
and
R.B.Honzatko
(2001).
Recombinant mouse muscle adenylosuccinate synthetase: overexpression, kinetics, and crystal structure.
|
| |
J Biol Chem,
276,
42146-42152.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
R.A.Farley,
E.Elquza,
J.Müller-Ehmsen,
D.J.Kane,
A.K.Nagy,
V.N.Kasho,
and
L.D.Faller
(2001).
18O-exchange evidence that mutations of arginine in a signature sequence for P-type pumps affect inorganic phosphate binding.
|
| |
Biochemistry,
40,
6361-6370.
|
 |
|
|
|
|
 |
J.B.Thoden,
F.M.Raushel,
G.Wesenberg,
and
H.M.Holden
(1999).
The binding of inosine monophosphate to Escherichia coli carbamoyl phosphate synthetase.
|
| |
J Biol Chem,
274,
22502-22507.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
J.Y.Choe,
B.W.Poland,
H.J.Fromm,
and
R.B.Honzatko
(1999).
Mechanistic implications from crystalline complexes of wild-type and mutant adenylosuccinate synthetases from Escherichia coli.
|
| |
Biochemistry,
38,
6953-6961.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
P.Lee,
A.Gorrell,
H.J.Fromm,
and
R.F.Colman
(1999).
Implication of arginine-131 and arginine-303 in the substrate site of adenylosuccinate synthetase of Escherichia coli by affinity labeling with 6-(4-bromo-2,3-dioxobutyl)thioadenosine 5'-monophosphate.
|
| |
Biochemistry,
38,
5754-5763.
|
 |
|
|
|
|
 |
Z.Hou,
M.Cashel,
H.J.Fromm,
and
R.B.Honzatko
(1999).
Effectors of the stringent response target the active site of Escherichia coli adenylosuccinate synthetase.
|
| |
J Biol Chem,
274,
17505-17510.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
C.Zeng,
A.E.Aleshin,
G.Chen,
R.B.Honzatko,
and
H.J.Fromm
(1998).
The roles of glycine residues in the ATP binding site of human brain hexokinase.
|
| |
J Biol Chem,
273,
700-704.
|
 |
|
|
|
|
 |
J.Y.Choe,
B.W.Poland,
H.J.Fromm,
and
R.B.Honzatko
(1998).
Role of a dynamic loop in cation activation and allosteric regulation of recombinant porcine fructose-1,6-bisphosphatase.
|
| |
Biochemistry,
37,
11441-11450.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
W.Wang,
A.Gorrell,
Z.Hou,
R.B.Honzatko,
and
H.J.Fromm
(1998).
Ambiguities in mapping the active site of a conformationally dynamic enzyme by directed mutation. Role of dynamics in structure-function correlations in Escherichia coli adenylosuccinate synthetase.
|
| |
J Biol Chem,
273,
16000-16004.
|
 |
|
|
|
|
 |
B.W.Poland,
C.Bruns,
H.J.Fromm,
and
R.B.Honzatko
(1997).
Entrapment of 6-thiophosphoryl-IMP in the active site of crystalline adenylosuccinate synthetase from Escherichia coli.
|
| |
J Biol Chem,
272,
15200-15205.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
W.Wang,
Z.Hou,
R.B.Honzatko,
and
H.J.Fromm
(1997).
Relationship of conserved residues in the IMP binding site to substrate recognition and catalysis in Escherichia coli adenylosuccinate synthetase.
|
| |
J Biol Chem,
272,
16911-16916.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

