 |
PDBsum entry 1loo

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.6.3.4.4
- adenylosuccinate synthase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
AMP and GMP Biosynthesis
|
 |
 |
 |
 |
 |
Reaction:
|
 |
IMP + L-aspartate + GTP = N6-(1,2-dicarboxyethyl)-AMP + GDP + phosphate + 2 H+
|
 |
 |
 |
 |
 |
IMP
Bound ligand (Het Group name = )
corresponds exactly
|
+
|
 L-aspartate
L-aspartate
|
+
|
 GTP
GTP
|
=
|
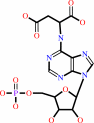 N(6)-(1,2-dicarboxyethyl)-AMP
N(6)-(1,2-dicarboxyethyl)-AMP
|
+
|
 GDP
GDP
|
+
|
 phosphate
phosphate
|
+
|
2
×
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Biol Chem
277:26779-26787
(2002)
|
|
PubMed id:
|
|
|
|

|
| |
|
IMP, GTP, and 6-phosphoryl-IMP complexes of recombinant mouse muscle adenylosuccinate synthetase.
|
|
C.V.Iancu,
T.Borza,
H.J.Fromm,
R.B.Honzatko.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Prokaryotes have a single form of adenylosuccinate synthetase that controls the
committed step of AMP biosynthesis, but vertebrates have two isozymes of the
synthetase. The basic isozyme, which predominates in muscle, participates in the
purine nucleotide cycle, has an active site conformation different from that of
the Escherichia coli enzyme, and exhibits significant differences in ligand
recognition. Crystalline complexes presented here of the recombinant basic
isozyme from mouse show the following. GTP alone binds to the active site
without inducing a conformational change. IMP in combination with an acetate
anion induces major conformational changes and organizes the active site for
catalysis. IMP, in the absence of GTP, binds to the GTP pocket of the
synthetase. The combination of GTP and IMP results in the formation of a stable
complex of 6-phosphoryl-IMP and GDP in the presence or absence of hadacidin. The
response of the basic isozyme to GTP alone differs from that of synthetases from
plants, and yet the conformation of the mouse basic and E. coli synthetases in
their complexes with GDP, 6-phosphoryl-IMP, and hadacidin are nearly identical.
Hence, reported differences in ligand recognition among synthetases probably
arise from conformational variations observed in partially ligated enzymes.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 2.
Fig. 2. Stereoview of GTP bound to the active site.
Electron density is from an omit map (coefficients of 2F[obs]
 F[calc], F[calc],
 [calc]
phases) contoured at 3 [calc]
phases) contoured at 3  using a
cutoff radius of 1 Å. Dashed lines represent
donor-acceptor interactions. using a
cutoff radius of 1 Å. Dashed lines represent
donor-acceptor interactions.
|
 |
Figure 6.
Fig. 6. Stereoview of the
6PIMP·GDP·hadacidin complex. One molecule of
6PIMP, GDP, and hadacidin bind to the single, symmetry-unique
subunit in this crystal form ( top). Electron density is from an
omit map (coefficients of 2F[obs]  F[calc], F[calc],
 [calc]
phases) contoured at 3 [calc]
phases) contoured at 3  using a
cutoff radius of 1 Å. Dashed lines represent
donor-acceptor interactions. Only the new interactions, relative
to GDP·6PIMP complex, are shown. Additional details are
in "Results." using a
cutoff radius of 1 Å. Dashed lines represent
donor-acceptor interactions. Only the new interactions, relative
to GDP·6PIMP complex, are shown. Additional details are
in "Results."
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the ASBMB:
J Biol Chem
(2002,
277,
26779-26787)
copyright 2002.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
K.S.Chakrabarti,
K.G.Thakur,
B.Gopal,
and
S.P.Sarma
(2010).
X-ray crystallographic and NMR studies of pantothenate synthetase provide insights into the mechanism of homotropic inhibition by pantoate.
|
| |
FEBS J,
277,
697-712.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
E.R.Jefferson,
T.P.Walsh,
and
G.J.Barton
(2008).
A comparison of SCOP and CATH with respect to domain-domain interactions.
|
| |
Proteins,
70,
54-62.
|
 |
|
|
|
|
 |
F.R.Fischer,
P.A.Wood,
F.H.Allen,
and
F.Diederich
(2008).
Orthogonal dipolar interactions between amide carbonyl groups.
|
| |
Proc Natl Acad Sci U S A,
105,
17290-17294.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

