 |
PDBsum entry 1fq0

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.4.1.2.14
- 2-dehydro-3-deoxy-phosphogluconate aldolase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
2-dehydro-3-deoxy-6-phospho-D-gluconate = D-glyceraldehyde 3-phosphate + pyruvate
|
 |
 |
 |
 |
 |
 2-dehydro-3-deoxy-6-phospho-D-gluconate
2-dehydro-3-deoxy-6-phospho-D-gluconate
|
=
|
 D-glyceraldehyde 3-phosphate
D-glyceraldehyde 3-phosphate
|
+
|
pyruvate
Bound ligand (Het Group name = )
matches with 46.15% similarity
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 3:
|
 |
E.C.4.1.3.16
- 4-hydroxy-2-oxoglutarate aldolase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
|
1.
|
(4R)-4-hydroxy-2-oxoglutarate = glyoxylate + pyruvate
|
|
2.
|
(4S)-4-hydroxy-2-oxoglutarate = glyoxylate + pyruvate
|
|
 |
 |
 |
 |
 |
(4R)-4-hydroxy-2-oxoglutarate
|
=
|
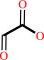 glyoxylate
glyoxylate
|
+
|
pyruvate
Bound ligand (Het Group name = )
matches with 46.15% similarity
|
|
 |
 |
 |
 |
 |
(4S)-4-hydroxy-2-oxoglutarate
|
=
|
 glyoxylate
glyoxylate
|
+
|
pyruvate
Bound ligand (Het Group name = )
matches with 46.15% similarity
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Structure
9:1-9
(2001)
|
|
PubMed id:
|
|
|
|

|
| |
|
Directed evolution of a new catalytic site in 2-keto-3-deoxy-6-phosphogluconate aldolase from Escherichia coli.
|
|
N.Wymer,
L.V.Buchanan,
D.Henderson,
N.Mehta,
C.H.Botting,
L.Pocivavsek,
C.A.Fierke,
E.J.Toone,
J.H.Naismith.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
BACKGROUND: Aldolases are carbon bond-forming enzymes that have long been
identified as useful tools for the organic chemist. However, their utility is
limited in part by their narrow substrate utilization. Site-directed mutagenesis
of various enzymes to alter their specificity has been performed for many years,
typically without the desired effect. More recently directed evolution has been
employed to engineer new activities onto existing scaffoldings. This approach
allows random mutation of the gene and then selects for fitness to purpose those
proteins with the desired activity. To date such approaches have furnished novel
activities through multiple mutations of residues involved in recognition; in no
instance has a key catalytic residue been altered while activity is retained.
RESULTS: We report a double mutant of E. coli 2-keto-3-deoxy-6-phosphogluconate
aldolase with reduced but measurable enzyme activity and a synthetically useful
substrate profile. The mutant was identified from directed-evolution
experiments. Modification of substrate specificity is achieved by altering the
position of the active site lysine from one beta strand to a neighboring strand
rather than by modification of the substrate recognition site. The new enzyme is
different to all other existing aldolases with respect to the location of its
active site to secondary structure. The new enzyme still displays enantiofacial
discrimination during aldol addition. We have determined the crystal structure
of the wild-type enzyme (by multiple wavelength methods) to 2.17 A and the
double mutant enzyme to 2.7 A resolution. CONCLUSIONS: These results suggest
that the scope of directed evolution is substantially larger than previously
envisioned in that it is possible to perturb the active site residues themselves
as well as surrounding loops to alter specificity. The structure of the double
mutant shows how catalytic competency is maintained despite spatial
reorganization of the active site with respect to substrate.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |
Figure 1.
Figure 1. The Reaction Catalyzed by KDPG Aldolase

|
 |
|
|

|
| |
The above figure is
reprinted
by permission from Cell Press:
Structure
(2001,
9,
1-9)
copyright 2001.
|
|
| |
Figure was
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
I.Campeotto,
S.B.Carr,
C.H.Trinh,
A.S.Nelson,
A.Berry,
S.E.Phillips,
and
A.R.Pearson
(2009).
Structure of an Escherichia coli N-acetyl-D-neuraminic acid lyase mutant, E192N, in complex with pyruvate at 1.45 angstrom resolution.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
65,
1088-1090.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
M.Pavlova,
M.Klvana,
Z.Prokop,
R.Chaloupkova,
P.Banas,
M.Otyepka,
R.C.Wade,
M.Tsuda,
Y.Nagata,
and
J.Damborsky
(2009).
Redesigning dehalogenase access tunnels as a strategy for degrading an anthropogenic substrate.
|
| |
Nat Chem Biol,
5,
727-733.
|
 |
|
|
|
|
 |
M.Cheriyan,
E.J.Toone,
and
C.A.Fierke
(2007).
Mutagenesis of the phosphate-binding pocket of KDPG aldolase enhances selectivity for hydrophobic substrates.
|
| |
Protein Sci,
16,
2368-2377.
|
 |
|
|
|
|
 |
A.K.Samland,
and
G.A.Sprenger
(2006).
Microbial aldolases as C-C bonding enzymes--unknown treasures and new developments.
|
| |
Appl Microbiol Biotechnol,
71,
253-264.
|
 |
|
|
|
|
 |
F.P.Seebeck,
A.Guainazzi,
C.Amoreira,
K.K.Baldridge,
and
D.Hilvert
(2006).
Stereoselectivity and expanded substrate scope of an engineered PLP-dependent aldolase.
|
| |
Angew Chem Int Ed Engl,
45,
6824-6826.
|
 |
|
|
|
|
 |
J.Kaur,
and
R.Sharma
(2006).
Directed evolution: an approach to engineer enzymes.
|
| |
Crit Rev Biotechnol,
26,
165-199.
|
 |
|
|
|
|
 |
B.Höcker
(2005).
Directed evolution of (betaalpha)(8)-barrel enzymes.
|
| |
Biomol Eng,
22,
31-38.
|
 |
|
|
|
|
 |
K.B.Murray,
W.R.Taylor,
and
J.M.Thornton
(2004).
Toward the detection and validation of repeats in protein structure.
|
| |
Proteins,
57,
365-380.
|
 |
|
|
|
|
 |
R.Fischetti,
S.Stepanov,
G.Rosenbaum,
R.Barrea,
E.Black,
D.Gore,
R.Heurich,
E.Kondrashkina,
A.J.Kropf,
S.Wang,
K.Zhang,
T.C.Irving,
and
G.B.Bunker
(2004).
The BioCAT undulator beamline 18ID: a facility for biological non-crystalline diffraction and X-ray absorption spectroscopy at the Advanced Photon Source.
|
| |
J Synchrotron Radiat,
11,
399-405.
|
 |
|
|
|
|
 |
R.H.Lilien,
C.Bailey-Kellogg,
A.C.Anderson,
and
B.R.Donald
(2004).
A subgroup algorithm to identify cross-rotation peaks consistent with non-crystallographic symmetry.
|
| |
Acta Crystallogr D Biol Crystallogr,
60,
1057-1067.
|
 |
|
|
|
|
 |
B.J.Bell,
L.Watanabe,
J.L.Rios-Steiner,
A.Tulinsky,
L.Lebioda,
and
R.K.Arni
(2003).
Structure of 2-keto-3-deoxy-6-phosphogluconate (KDPG) aldolase from Pseudomonas putida.
|
| |
Acta Crystallogr D Biol Crystallogr,
59,
1454-1458.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
E.J.de Vries,
and
D.B.Janssen
(2003).
Biocatalytic conversion of epoxides.
|
| |
Curr Opin Biotechnol,
14,
414-420.
|
 |
|
|
|
|
 |
A.E.Todd,
C.A.Orengo,
and
J.M.Thornton
(2002).
Plasticity of enzyme active sites.
|
| |
Trends Biochem Sci,
27,
419-426.
|
 |
|
|
|
|
 |
D.J.Brockwell,
G.S.Beddard,
J.Clarkson,
R.C.Zinober,
A.W.Blake,
J.Trinick,
P.D.Olmsted,
D.A.Smith,
and
S.E.Radford
(2002).
The effect of core destabilization on the mechanical resistance of I27.
|
| |
Biophys J,
83,
458-472.
|
 |
|
|
|
|
 |
H.Zhao,
K.Chockalingam,
and
Z.Chen
(2002).
Directed evolution of enzymes and pathways for industrial biocatalysis.
|
| |
Curr Opin Biotechnol,
13,
104-110.
|
 |
|
|
|
|
 |
M.Kroemer,
and
G.E.Schulz
(2002).
The structure of L-rhamnulose-1-phosphate aldolase (class II) solved by low-resolution SIR phasing and 20-fold NCS averaging.
|
| |
Acta Crystallogr D Biol Crystallogr,
58,
824-832.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
A.Heine,
G.DeSantis,
J.G.Luz,
M.Mitchell,
C.H.Wong,
and
I.A.Wilson
(2001).
Observation of covalent intermediates in an enzyme mechanism at atomic resolution.
|
| |
Science,
294,
369-374.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
E.T.Farinas,
T.Bulter,
and
F.H.Arnold
(2001).
Directed enzyme evolution.
|
| |
Curr Opin Biotechnol,
12,
545-551.
|
 |
|
|
|
|
 |
W.D.Fessner,
and
V.Helaine
(2001).
Biocatalytic synthesis of hydroxylated natural products using aldolases and related enzymes.
|
| |
Curr Opin Biotechnol,
12,
574-586.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

