 |
PDBsum entry 1jcl

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.4.1.2.4
- deoxyribose-phosphate aldolase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Deoxyribose-phosphate aldolase
|
 |
 |
 |
 |
 |
Reaction:
|
 |
2-deoxy-D-ribose 5-phosphate = D-glyceraldehyde 3-phosphate + acetaldehyde
|
 |
 |
 |
 |
 |
2-deoxy-D-ribose 5-phosphate
Bound ligand (Het Group name = )
corresponds exactly
|
=
|
 D-glyceraldehyde 3-phosphate
D-glyceraldehyde 3-phosphate
|
+
|
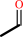 acetaldehyde
acetaldehyde
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Science
294:369-374
(2001)
|
|
PubMed id:
|
|
|
|

|
| |
|
Observation of covalent intermediates in an enzyme mechanism at atomic resolution.
|
|
A.Heine,
G.DeSantis,
J.G.Luz,
M.Mitchell,
C.H.Wong,
I.A.Wilson.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
In classical enzymology, intermediates and transition states in a catalytic
mechanism are usually inferred from a series of biochemical experiments. Here,
we derive an enzyme mechanism from true atomic-resolution x-ray structures of
reaction intermediates. Two ultra-high resolution structures of wild-type and
mutant d-2-deoxyribose-5-phosphate (DRP) aldolase complexes with DRP at 1.05 and
1.10 angstroms unambiguously identify the postulated covalent carbinolamine and
Schiff base intermediates in the aldolase mechanism. In combination with
site-directed mutagenesis and (1)H nuclear magnetic resonance, we can now
propose how the heretofore elusive C-2 proton abstraction step and the overall
stereochemical course are accomplished. A proton relay system appears to
activate a conserved active-site water that functions as the critical mediator
for proton transfer.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
Fig. 1. DERA reaction. In vivo, DERA catalyzes the aldol
reaction between the acetaldehyde donor and
D-glyceraldehyde-3-phosphate acceptor to generate DRP.
|
 |
Figure 4.
Fig. 4. Catalytic mechanism for DERA. (A) 1H NMR experiment.
DERA-catalyzed exchange of C2 proton of
(R)-2-deuteropropanaldehyde and (S)-2-deuteropropanaldehyde in
D[2]O was established by 1H NMR. Incubation of
(R)-2-deuteropropanaldehyde with DERA in D[2]O effects complete
exchange of the  -proton to
a deuteron, as observed by the collapse of the C3 doublet
resonances of the aldehyde and acetal to singlets (left). By
contrast, incubation of (S)-2-deuteropropanaldehyde with DERA in
D[2]O does not effect proton exchange, as observed by retention
of the doublet resonances (right). (B) Proposed catalytic
mechanism for DERA. The proposed mechanism is consistent with
all of our ultra-high resolution structural, modeling,
site-directed mutagenesis, and 1H NMR data. Lys167 is identified
as the Schiff base-forming residue. After the enamine is formed,
the system is poised for nucleophilic attack onto the
carbonyl-carbon of the acceptor aldehyde
D-glyceraldehyde-3-phosphate. A proton relay system between
Asp102, Lys201, and an active site water molecule is responsible
for shuffling a proton between C2 of the acetaldehyde imine and
enamine and subsequent C3 hydroxyl protonation. The double
arrows in green indicate rapid proton shuffling between Lys201
and Asp102. Crystallographically observed reaction intermediates
are boxed (left, carbinolamine; right, Schiff base). (C)
Stereochemical course of aldol reaction. When propanal is
substituted for acetaldehyde as the donor, the pro-S -proton to
a deuteron, as observed by the collapse of the C3 doublet
resonances of the aldehyde and acetal to singlets (left). By
contrast, incubation of (S)-2-deuteropropanaldehyde with DERA in
D[2]O does not effect proton exchange, as observed by retention
of the doublet resonances (right). (B) Proposed catalytic
mechanism for DERA. The proposed mechanism is consistent with
all of our ultra-high resolution structural, modeling,
site-directed mutagenesis, and 1H NMR data. Lys167 is identified
as the Schiff base-forming residue. After the enamine is formed,
the system is poised for nucleophilic attack onto the
carbonyl-carbon of the acceptor aldehyde
D-glyceraldehyde-3-phosphate. A proton relay system between
Asp102, Lys201, and an active site water molecule is responsible
for shuffling a proton between C2 of the acetaldehyde imine and
enamine and subsequent C3 hydroxyl protonation. The double
arrows in green indicate rapid proton shuffling between Lys201
and Asp102. Crystallographically observed reaction intermediates
are boxed (left, carbinolamine; right, Schiff base). (C)
Stereochemical course of aldol reaction. When propanal is
substituted for acetaldehyde as the donor, the pro-S  proton is
removed, and thus the aldol reaction proceeds with retention of
configuration at C2, with the Si face of the resulting enamine
approaching the Re face of the acceptor carbonyl. A:H, general
acid; B, general base. proton is
removed, and thus the aldol reaction proceeds with retention of
configuration at C2, with the Si face of the resulting enamine
approaching the Re face of the acceptor carbonyl. A:H, general
acid; B, general base.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the AAAs:
Science
(2001,
294,
369-374)
copyright 2001.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
A.Fihri,
D.Luart,
C.Len,
A.Solhy,
C.Chevrin,
and
V.Polshettiwar
(2011).
Suzuki-Miyaura cross-coupling coupling reactions with low catalyst loading: a green and sustainable protocol in pure water.
|
| |
Dalton Trans,
40,
3116-3121.
|
 |
|
|
|
|
 |
R.Shi,
L.McDonald,
Q.Cui,
A.Matte,
M.Cygler,
and
I.Ekiel
(2011).
Structural and mechanistic insight into covalent substrate binding by Escherichia coli dihydroxyacetone kinase.
|
| |
Proc Natl Acad Sci U S A,
108,
1302-1307.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
H.H.Nguyen,
L.Wang,
H.Huang,
E.Peisach,
D.Dunaway-Mariano,
and
K.N.Allen
(2010).
Structural determinants of substrate recognition in the HAD superfamily member D-glycero-D-manno-heptose-1,7-bisphosphate phosphatase (GmhB) .
|
| |
Biochemistry,
49,
1082-1092.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
A.S.Burton,
and
N.Lehman
(2009).
DNA before proteins? Recent discoveries in nucleic acid catalysis strengthen the case.
|
| |
Astrobiology,
9,
125-130.
|
 |
|
|
|
|
 |
E.W.Debler,
R.Müller,
D.Hilvert,
and
I.A.Wilson
(2009).
An aspartate and a water molecule mediate efficient acid-base catalysis in a tailored antibody pocket.
|
| |
Proc Natl Acad Sci U S A,
106,
18539-18544.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
H.Huang,
H.Y.Kim,
I.D.Kozekov,
Y.J.Cho,
H.Wang,
A.Kozekova,
T.M.Harris,
C.J.Rizzo,
and
M.P.Stone
(2009).
Stereospecific formation of the (R)-gamma-hydroxytrimethylene interstrand N2-dG:N2-dG cross-link arising from the gamma-OH-1,N2-propano-2'-deoxyguanosine adduct in the 5'-CpG-3' DNA sequence.
|
| |
J Am Chem Soc,
131,
8416-8424.
|
 |
|
|
|
|
 |
J.A.Gerlt,
and
P.C.Babbitt
(2009).
Enzyme (re)design: lessons from natural evolution and computation.
|
| |
Curr Opin Chem Biol,
13,
10-18.
|
 |
|
|
|
|
 |
L.W.Xu,
J.Luo,
and
Y.Lu
(2009).
Asymmetric catalysis with chiral primary amine-based organocatalysts.
|
| |
Chem Commun (Camb),
(),
1807-1821.
|
 |
|
|
|
|
 |
M.M.Müller,
M.A.Windsor,
W.C.Pomerantz,
S.H.Gellman,
and
D.Hilvert
(2009).
A rationally designed aldolase foldamer.
|
| |
Angew Chem Int Ed Engl,
48,
922-925.
|
 |
|
|
|
|
 |
M.Raj,
and
V.K.Singh
(2009).
Organocatalytic reactions in water.
|
| |
Chem Commun (Camb),
(),
6687-6703.
|
 |
|
|
|
|
 |
T.Kawamichi,
T.Haneda,
M.Kawano,
and
M.Fujita
(2009).
X-ray observation of a transient hemiaminal trapped in a porous network.
|
| |
Nature,
461,
633-635.
|
 |
|
|
|
|
 |
Z.Diaz,
K.B.Xavier,
and
S.T.Miller
(2009).
The crystal structure of the Escherichia coli autoinducer-2 processing protein LsrF.
|
| |
PLoS One,
4,
e6820.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
Z.Xu,
P.Daka,
and
H.Wang
(2009).
Primary amine-metal Lewis acid bifunctional catalysts: the application to asymmetric direct aldol reactions.
|
| |
Chem Commun (Camb),
(),
6825-6827.
|
 |
|
|
|
|
 |
D.S.Berkholz,
H.R.Faber,
S.N.Savvides,
and
P.A.Karplus
(2008).
Catalytic cycle of human glutathione reductase near 1 A resolution.
|
| |
J Mol Biol,
382,
371-384.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
L.Jiang,
E.A.Althoff,
F.R.Clemente,
L.Doyle,
D.Röthlisberger,
A.Zanghellini,
J.L.Gallaher,
J.L.Betker,
F.Tanaka,
C.F.Barbas,
D.Hilvert,
K.N.Houk,
B.L.Stoddard,
and
D.Baker
(2008).
De novo computational design of retro-aldol enzymes.
|
| |
Science,
319,
1387-1391.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
L.W.Xu,
and
Y.Lu
(2008).
Primary amino acids: privileged catalysts in enantioselective organocatalysis.
|
| |
Org Biomol Chem,
6,
2047-2053.
|
 |
|
|
|
|
 |
H.Sakuraba,
K.Yoneda,
K.Yoshihara,
K.Satoh,
R.Kawakami,
Y.Uto,
H.Tsuge,
K.Takahashi,
H.Hori,
and
T.Ohshima
(2007).
Sequential aldol condensation catalyzed by hyperthermophilic 2-deoxy-d-ribose-5-phosphate aldolase.
|
| |
Appl Environ Microbiol,
73,
7427-7434.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
J.Dechancie,
F.R.Clemente,
A.J.Smith,
H.Gunaydin,
Y.L.Zhao,
X.Zhang,
and
K.N.Houk
(2007).
How similar are enzyme active site geometries derived from quantum mechanical theozymes to crystal structures of enzyme-inhibitor complexes? Implications for enzyme design.
|
| |
Protein Sci,
16,
1851-1866.
|
 |
|
|
|
|
 |
T.Iwasawa,
R.J.Hooley,
and
J.Rebek
(2007).
Stabilization of labile carbonyl addition intermediates by a synthetic receptor.
|
| |
Science,
317,
493-496.
|
 |
|
|
|
|
 |
Y.L.Boersma,
M.J.Dröge,
and
W.J.Quax
(2007).
Selection strategies for improved biocatalysts.
|
| |
FEBS J,
274,
2181-2195.
|
 |
|
|
|
|
 |
A.Córdova,
W.Zou,
P.Dziedzic,
I.Ibrahem,
E.Reyes,
and
Y.Xu
(2006).
Direct asymmetric intermolecular aldol reactions catalyzed by amino acids and small peptides.
|
| |
Chemistry,
12,
5383-5397.
|
 |
|
|
|
|
 |
P.Dziedzic,
W.Zou,
J.Háfren,
and
A.Córdova
(2006).
The small peptide-catalyzed direct asymmetric aldol reaction in water.
|
| |
Org Biomol Chem,
4,
38-40.
|
 |
|
|
|
|
 |
S.Jennewein,
M.Schürmann,
M.Wolberg,
I.Hilker,
R.Luiten,
M.Wubbolts,
and
D.Mink
(2006).
Directed evolution of an industrial biocatalyst: 2-deoxy-D-ribose 5-phosphate aldolase.
|
| |
Biotechnol J,
1,
537-548.
|
 |
|
|
|
|
 |
A.Bassan,
W.Zou,
E.Reyes,
F.Himo,
and
A.Córdova
(2005).
The origin of stereoselectivity in primary amino acid catalyzed intermolecular aldol reactions.
|
| |
Angew Chem Int Ed Engl,
44,
7028-7032.
|
 |
|
|
|
|
 |
A.Córdova,
I.Ibrahem,
J.Casas,
H.Sundén,
M.Engqvist,
and
E.Reyes
(2005).
Amino acid catalyzed neogenesis of carbohydrates: a plausible ancient transformation.
|
| |
Chemistry,
11,
4772-4784.
|
 |
|
|
|
|
 |
C.C.Hsu,
Z.Hong,
M.Wada,
D.Franke,
and
C.H.Wong
(2005).
Directed evolution of D-sialic acid aldolase to L-3-deoxy-manno-2-octulosonic acid (L-KDO) aldolase.
|
| |
Proc Natl Acad Sci U S A,
102,
9122-9126.
|
 |
|
|
|
|
 |
J.Casas,
M.Engqvist,
I.Ibrahem,
B.Kaynak,
and
A.Córdova
(2005).
Direct amino acid catalyzed asymmetric synthesis of polyketide sugars.
|
| |
Angew Chem Int Ed Engl,
44,
1343-1345.
|
 |
|
|
|
|
 |
K.Wang,
and
R.Samudrala
(2005).
FSSA: a novel method for identifying functional signatures from structural alignments.
|
| |
Bioinformatics,
21,
2969-2977.
|
 |
|
|
|
|
 |
M.Limbach
(2005).
'Five at one stroke': proline and small peptides in the stereoselective de novo synthesis and enantiotopic functionalization of carbohydrates.
|
| |
Chem Biodivers,
2,
825-836.
|
 |
|
|
|
|
 |
A.W.Schüttelkopf,
and
D.M.van Aalten
(2004).
PRODRG: a tool for high-throughput crystallography of protein-ligand complexes.
|
| |
Acta Crystallogr D Biol Crystallogr,
60,
1355-1363.
|
 |
|
|
|
|
 |
L.Yang,
J.S.Dordick,
and
S.Garde
(2004).
Hydration of enzyme in nonaqueous media is consistent with solvent dependence of its activity.
|
| |
Biophys J,
87,
812-821.
|
 |
|
|
|
|
 |
N.K.Lokanath,
I.Shiromizu,
N.Ohshima,
Y.Nodake,
M.Sugahara,
S.Yokoyama,
S.Kuramitsu,
M.Miyano,
and
N.Kunishima
(2004).
Structure of aldolase from Thermus thermophilus HB8 showing the contribution of oligomeric state to thermostability.
|
| |
Acta Crystallogr D Biol Crystallogr,
60,
1816-1823.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
N.Rashid,
H.Imanaka,
T.Fukui,
H.Atomi,
and
T.Imanaka
(2004).
Presence of a novel phosphopentomutase and a 2-deoxyribose 5-phosphate aldolase reveals a metabolic link between pentoses and central carbon metabolism in the hyperthermophilic archaeon Thermococcus kodakaraensis.
|
| |
J Bacteriol,
186,
4185-4191.
|
 |
|
|
|
|
 |
C.M.Wilmot,
and
A.R.Pearson
(2002).
Cryocrystallography of metalloprotein reaction intermediates.
|
| |
Curr Opin Chem Biol,
6,
202-207.
|
 |
|
|
|
|
 |
J.L.Hansen,
J.A.Ippolito,
N.Ban,
P.Nissen,
P.B.Moore,
and
T.A.Steitz
(2002).
The structures of four macrolide antibiotics bound to the large ribosomal subunit.
|
| |
Mol Cell,
10,
117-128.
|
 |
|
PDB codes:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

