
|

|

|
|
 |
Contents |
 |
|
|

|

|

|

|

|

|

|



 103 a.a.
103 a.a.
|
 |

|

|

|

|

|

|

|
 31 a.a.
31 a.a.
|
 |

|

|

|

|

|

|

|
 27 a.a.
27 a.a.
|
 |

|

|

|

|

|

|

|
 27 a.a.
27 a.a.
|
 |

|

|

|

|

|

|

|
 26 a.a.
26 a.a.
|
 |

|

|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Transcription
|
 |
|
Title:
|
 |
Crystal structure of a complex between the dimerization domain of hnf- 1 alpha and the coactivator dcoh
|
|
Structure:
|
 |
Dimerization cofactor of hepatocyte nuclear factor 1-alpha. Chain: a, b, c, d. Synonym: pterin-4-alpha-carbinolamine dehydratase, phs, dcoh. Engineered: yes. Hepatocyte nuclear factor 1-alpha. Chain: e, f, g, h. Fragment: dimerization domain (residues 1-32). Engineered: yes
|
|
Source:
|
 |
Rattus norvegicus. Norway rat. Organism_taxid: 10116. Organ: liver. Expressed in: escherichia coli. Expression_system_taxid: 562. Synthetic: yes. Other_details: this peptide was chemically synthesized. The sequence of this peptide naturally occurs in mouse (mus musculus)
|
|
Biol. unit:
|
 |
 Tetramer (from
)
Tetramer (from
)
|
|
Resolution:
|
 |
|
2.60Å
|
R-factor:
|
0.257
|
R-free:
|
0.299
|
|
|
Authors:
|
 |
R.B.Rose,J.H.Bayle,J.A.Endrizzi,J.D.Cronk,G.R.Crabtree,T.Alber
|
Key ref:

|
 |
R.B.Rose
et al.
(2000).
Structural basis of dimerization, coactivator recognition and MODY3 mutations in HNF-1alpha.
Nat Struct Biol,
7,
744-748.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
06-Jul-00
|
Release date:
|
20-Sep-00
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P61459
(PHS_RAT) -
Pterin-4-alpha-carbinolamine dehydratase from Rattus norvegicus
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
104 a.a.
103 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|

|

|
|
P22361
(HNF1A_MOUSE) -
Hepatocyte nuclear factor 1-alpha from Mus musculus
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
628 a.a.
31 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|

|

|
|
P22361
(HNF1A_MOUSE) -
Hepatocyte nuclear factor 1-alpha from Mus musculus
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
628 a.a.
27 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
Chains A, B, C, D:
E.C.4.2.1.96
- 4a-hydroxytetrahydrobiopterin dehydratase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Biopterin Biosynthesis
|
 |
 |
 |
 |
 |
Reaction:
|
 |
(4aS,6R)-4a-hydroxy-L-erythro-5,6,7,8-tetrahydrobiopterin = (6R)-L- erythro-6,7-dihydrobiopterin + H2O
|
 |
 |
 |
 |
 |
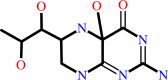 4a-hydroxytetrahydrobiopterin
4a-hydroxytetrahydrobiopterin
|
=
|
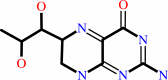 6,7-dihydrobiopterin
6,7-dihydrobiopterin
|
+
|
H(2)O
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Nat Struct Biol
7:744-748
(2000)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structural basis of dimerization, coactivator recognition and MODY3 mutations in HNF-1alpha.
|
|
R.B.Rose,
J.H.Bayle,
J.A.Endrizzi,
J.D.Cronk,
G.R.Crabtree,
T.Alber.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Maturity-onset diabetes of the young type 3 (MODY3) results from mutations in
the transcriptional activator hepatocyte nuclear factor-1alpha (HNF-1alpha).
Several MODY3 mutations target the HNF-1alpha dimerization domain (HNF-p1),
which binds the coactivator, dimerization cofactor of HNF-1 (DCoH). To define
the mechanism of coactivator recognition and the basis for the MODY3 phenotype,
we determined the cocrystal structure of the DCoH-HNF-p1 complex and
characterized biochemically the effects of MODY3 mutations in HNF-p1. The
DCoH-HNF-p1 complex comprises a dimer of dimers in which HNF-p1 forms a unique
four-helix bundle. Through rearrangements of interfacial side chains, a single,
bifunctional interface in the DCoH dimer mediates both HNF-1alpha binding and
formation of a competing, transcriptionally inactive DCoH homotetramer.
Consistent with the structure, MODY3 mutations in HNF-p1 reduce activator
function by two distinct mechanisms.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
Figure 1. Stereo view of the experimental, MAD-phased 2.6 Å
resolution, electron density map contoured at 1  superimposed
on the refined model. Residues 8 -11 in helix 1 of HNF-p1 and
the interacting amino acids of the DCoH dimer (residues 55 -57
and 43' -46', where primes denote residues in the neighboring
subunit) are shown. superimposed
on the refined model. Residues 8 -11 in helix 1 of HNF-p1 and
the interacting amino acids of the DCoH dimer (residues 55 -57
and 43' -46', where primes denote residues in the neighboring
subunit) are shown.
|
 |
Figure 2.
Figure 2. Structure of the DCoH -HNF-p1 complex. a, The DCoH
dimer (yellow and orange) binds the HNF-p1 dimer (light and dark
blue) with two helix 2 sequences of DCoH in contact with two
helix 1 sequences of HNF-p1. The two-fold rotation axes of the
dimers coincide (arrow). b, The bound HNF-p1 dimer (light and
dark blue) forms an antiparallel four-helix bundle. The view is
along the two-fold rotation axis (+) with the DCoH binding
surface in front. Seven Leu side chains in each monomer
stabilize the dimer and make contacts with DCoH. Red spheres
mark two residues mutated in MODY3 patients, Leu 12 and Gly 20.
The site of a third MODY3 mutation, Gly 31, occurs in the
disordered region of the chain beyond residue 30. c,
Electrostatic potential displayed on the surface of the
recognition helices of DCoH (red, <-2.5 kT/e; white, -2.5 to 2.5
kT/e; and blue, >2.5 kT/e). A stick representation of helix 1 of
the HNF-p1 dimer (light blue) is superimposed on the surface. d,
The corresponding electrostatic potential displayed on the
surface of the HNF-1  recognition
helices. A stick representation of the recognition helices of
the bound DCoH (yellow) is superimposed. The two surfaces in (c)
and (d) match through a 180° rotation about a central, vertical
axis. Complementary positive (DCoH) and negative (HNF-p1)
potentials are evident on the left and right edges of the two
interfaces. e, Interactions between DCoH (yellow and orange) and
HNF-p1 (light and dark blue), viewed along the DCoH helical
axes. Only half of each helix is shown, because the interactions
in the other half are identical. Hydrophobic residues forming
the core of the interface are displayed in gray. Side chains
within hydrogen bonding distance are connected by lines. DCoH
Glu 58' caps the N-terminus of the HNF-p1 helix 1, the Leu 8
amide, and forms a hydrogen bond with Ser 6. DCOH Lys 59' forms
a hydrogen bond to the C-terminus of the neighboring HNF-p1
helix, the carbonyl of Ser 19'. recognition
helices. A stick representation of the recognition helices of
the bound DCoH (yellow) is superimposed. The two surfaces in (c)
and (d) match through a 180° rotation about a central, vertical
axis. Complementary positive (DCoH) and negative (HNF-p1)
potentials are evident on the left and right edges of the two
interfaces. e, Interactions between DCoH (yellow and orange) and
HNF-p1 (light and dark blue), viewed along the DCoH helical
axes. Only half of each helix is shown, because the interactions
in the other half are identical. Hydrophobic residues forming
the core of the interface are displayed in gray. Side chains
within hydrogen bonding distance are connected by lines. DCoH
Glu 58' caps the N-terminus of the HNF-p1 helix 1, the Leu 8
amide, and forms a hydrogen bond with Ser 6. DCOH Lys 59' forms
a hydrogen bond to the C-terminus of the neighboring HNF-p1
helix, the carbonyl of Ser 19'.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from Macmillan Publishers Ltd:
Nat Struct Biol
(2000,
7,
744-748)
copyright 2000.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
T.Jin,
F.Guo,
I.G.Serebriiskii,
A.Howard,
and
Y.Z.Zhang
(2007).
A 1.55 A resolution X-ray crystal structure of HEF2/ERH and insights into its transcriptional and cell-cycle interaction networks.
|
| |
Proteins,
68,
427-437.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
D.Jung,
Y.Choi,
and
M.Uesugi
(2006).
Small organic molecules that modulate gene transcription.
|
| |
Drug Discov Today,
11,
452-457.
|
 |
|
|
|
|
 |
S.J.Demarest,
S.Deechongkit,
H.J.Dyson,
R.M.Evans,
and
P.E.Wright
(2004).
Packing, specificity, and mutability at the binding interface between the p160 coactivator and CREB-binding protein.
|
| |
Protein Sci,
13,
203-210.
|
 |
|
|
|
|
 |
A.J.Warren
(2002).
Eukaryotic transcription factors.
|
| |
Curr Opin Struct Biol,
12,
107-114.
|
 |
|
|
|
|
 |
J.H.Bayle,
F.Randazzo,
G.Johnen,
S.Kaufman,
A.Nagy,
J.Rossant,
and
G.R.Crabtree
(2002).
Hyperphenylalaninemia and impaired glucose tolerance in mice lacking the bifunctional DCoH gene.
|
| |
J Biol Chem,
277,
28884-28891.
|
 |
|
|
|
|
 |
Y.I.Chi,
J.D.Frantz,
B.C.Oh,
L.Hansen,
S.Dhe-Paganon,
and
S.E.Shoelson
(2002).
Diabetes mutations delineate an atypical POU domain in HNF-1alpha.
|
| |
Mol Cell,
10,
1129-1137.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
E.Pogge von Strandmann,
S.Senkel,
and
G.U.Ryffel
(2001).
ERH (enhancer of rudimentary homologue), a conserved factor identical between frog and human, is a transcriptional repressor.
|
| |
Biol Chem,
382,
1379-1385.
|
 |
|
|
|
|
 |
M.G.Newlon,
M.Roy,
D.Morikis,
D.W.Carr,
R.Westphal,
J.D.Scott,
and
P.A.Jennings
(2001).
A novel mechanism of PKA anchoring revealed by solution structures of anchoring complexes.
|
| |
EMBO J,
20,
1651-1662.
|
 |
|
PDB codes:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|

| |

