 |
PDBsum entry 1e5h

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
1e5h
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.14.20.1
- deacetoxycephalosporin-C synthase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Penicillin N and Deacetoxycephalosporin C Biosynthesis
|
 |
 |
 |
 |
 |
Reaction:
|
 |
penicillin N + 2-oxoglutarate + O2 = deacetoxycephalosporin C + succinate + CO2 + H2O
|
 |
 |
 |
 |
 |
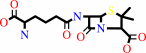 penicillin N
penicillin N
|
+
|
 2-oxoglutarate
2-oxoglutarate
|
+
|
 O2
O2
|
=
|
deacetoxycephalosporin C
Bound ligand (Het Group name = )
corresponds exactly
|
+
|
 succinate
succinate
|
+
|
 CO2
CO2
|
+
|
H2O
Bound ligand (Het Group name = )
corresponds exactly
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Mol Biol
308:937-948
(2001)
|
|
PubMed id:
|
|
|
|

|
| |
|
Kinetic and crystallographic studies on deacetoxycephalosporin C synthase (DAOCS).
|
|
H.J.Lee,
M.D.Lloyd,
K.Harlos,
I.J.Clifton,
J.E.Baldwin,
C.J.Schofield.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Deacetoxycephalosporin C synthase (DAOCS) is an iron(II) and
2-oxoglutarate-dependent oxygenase that catalyzes the conversion of penicillin N
to deacetoxycephalosporin C, the committed step in the biosynthesis of
cephalosporin antibiotics. The crystal structure of DAOCS revealed that the C
terminus of one molecule is inserted into the active site of its neighbor in a
cyclical fashion within a trimeric unit. This arrangement has hindered the
generation of crystalline enzyme-substrate complexes. Therefore, we constructed
a series of DAOCS mutants with modified C termini. Oxidation of 2-oxoglutarate
was significantly uncoupled from oxidation of the penicillin substrate in
certain truncated mutants. The extent of uncoupling varied with the number of
residues deleted and the penicillin substrate used. Crystal structures were
determined for the DeltaR306 mutant complexed with iron(II) and 2-oxoglutarate
(to 2.10 A) and the DeltaR306A mutant complexed with iron(II), succinate and
unhydrated carbon dioxide (to 1.96 A). The latter may mimic a product complex,
and supports proposals for a metal-bound CO(2) intermediate during catalysis.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
Figure 1. Structure of (a) iron(II) and 2-oxoglutarate
complex of wild-type enzyme [Valegard et al 1998]; (b) iron(II)
and 2-oxoglutarate complex of the DR306 mutant; and (c)
iron(II), succinate and CO[2] complex of the DR307A mutant. The
jelly-roll motif is shown in red, C termini are shown in blue,
and the C terminus from symmetry related molecules in green.
|
 |
Figure 3.
Figure 3. A view of the structure of the DR307A mutant
iron(II), succinate and CO[2] complex. (a) A similar view to
that in Figure 2, with the above residues omitted for clarity.
The electron density map is contoured to 1.29s; (b) close up
view of the CO[2]-binding site with succinate and the
side-chains of Val262 and Phe264 enclosing one face. The
remaining face of the molecule is bordered by residues in the C
terminus, the conformation of which may be significantly
different in solution (see the text for a discussion). The
electron density map is contoured to 1.69s.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from Elsevier:
J Mol Biol
(2001,
308,
937-948)
copyright 2001.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
R.Chowdhury,
K.K.Yeoh,
Y.M.Tian,
L.Hillringhaus,
E.A.Bagg,
N.R.Rose,
I.K.Leung,
X.S.Li,
E.C.Woon,
M.Yang,
M.A.McDonough,
O.N.King,
I.J.Clifton,
R.J.Klose,
T.D.Claridge,
P.J.Ratcliffe,
C.J.Schofield,
and
A.Kawamura
(2011).
The oncometabolite 2-hydroxyglutarate inhibits histone lysine demethylases.
|
| |
EMBO Rep,
12,
463-469.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
X.B.Wu,
X.Y.Tian,
J.J.Ji,
W.B.Wu,
K.Q.Fan,
and
K.Q.Yang
(2011).
Saturation mutagenesis of Acremonium chrysogenum deacetoxy/deacetylcephalosporin C synthase R308 site confirms its role in controlling substrate specificity.
|
| |
Biotechnol Lett,
33,
805-812.
|
 |
|
|
|
|
 |
E.Flashman,
E.A.Bagg,
R.Chowdhury,
J.Mecinović,
C.Loenarz,
M.A.McDonough,
K.S.Hewitson,
and
C.J.Schofield
(2008).
Kinetic rationale for selectivity toward N- and C-terminal oxygen-dependent degradation domain substrates mediated by a loop region of hypoxia-inducible factor prolyl hydroxylases.
|
| |
J Biol Chem,
283,
3808-3815.
|
 |
|
|
|
|
 |
J.J.Cotelesage,
J.Puttick,
H.Goldie,
B.Rajabi,
B.Novakovski,
and
L.T.Delbaere
(2007).
How does an enzyme recognize CO2?
|
| |
Int J Biochem Cell Biol,
39,
1204-1210.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
K.S.Hewitson,
B.M.Liénard,
M.A.McDonough,
I.J.Clifton,
D.Butler,
A.S.Soares,
N.J.Oldham,
L.A.McNeill,
and
C.J.Schofield
(2007).
Structural and mechanistic studies on the inhibition of the hypoxia-inducible transcription factor hydroxylases by tricarboxylic acid cycle intermediates.
|
| |
J Biol Chem,
282,
3293-3301.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
V.Purpero,
and
G.R.Moran
(2007).
The diverse and pervasive chemistries of the alpha-keto acid dependent enzymes.
|
| |
J Biol Inorg Chem,
12,
587-601.
|
 |
|
|
|
|
 |
I.Castro-Rodríguez,
and
K.Meyer
(2006).
Small molecule activation at uranium coordination complexes: control of reactivity via molecular architecture.
|
| |
Chem Commun (Camb),
(),
1353-1368.
|
 |
|
|
|
|
 |
M.A.McDonough,
V.Li,
E.Flashman,
R.Chowdhury,
C.Mohr,
B.M.Liénard,
J.Zondlo,
N.J.Oldham,
I.J.Clifton,
J.Lewis,
L.A.McNeill,
R.J.Kurzeja,
K.S.Hewitson,
E.Yang,
S.Jordan,
R.S.Syed,
and
C.J.Schofield
(2006).
Cellular oxygen sensing: Crystal structure of hypoxia-inducible factor prolyl hydroxylase (PHD2).
|
| |
Proc Natl Acad Sci U S A,
103,
9814-9819.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.L.Neidig,
A.Decker,
O.W.Choroba,
F.Huang,
M.Kavana,
G.R.Moran,
J.B.Spencer,
and
E.I.Solomon
(2006).
Spectroscopic and electronic structure studies of aromatic electrophilic attack and hydrogen-atom abstraction by non-heme iron enzymes.
|
| |
Proc Natl Acad Sci U S A,
103,
12966-12973.
|
 |
|
|
|
|
 |
Y.Mishina,
and
C.He
(2006).
Oxidative dealkylation DNA repair mediated by the mononuclear non-heme iron AlkB proteins.
|
| |
J Inorg Biochem,
100,
670-678.
|
 |
|
|
|
|
 |
Y.Mishina,
E.M.Duguid,
and
C.He
(2006).
Direct reversal of DNA alkylation damage.
|
| |
Chem Rev,
106,
215-232.
|
 |
|
|
|
|
 |
C.L.Wei,
Y.B.Yang,
C.H.Deng,
W.C.Liu,
J.S.Hsu,
Y.C.Lin,
S.H.Liaw,
and
Y.C.Tsai
(2005).
Directed evolution of Streptomyces clavuligerus deacetoxycephalosporin C synthase for enhancement of penicillin G expansion.
|
| |
Appl Environ Microbiol,
71,
8873-8880.
|
 |
|
|
|
|
 |
M.A.McDonough,
K.L.Kavanagh,
D.Butler,
T.Searls,
U.Oppermann,
and
C.J.Schofield
(2005).
Structure of human phytanoyl-CoA 2-hydroxylase identifies molecular mechanisms of Refsum disease.
|
| |
J Biol Chem,
280,
41101-41110.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
M.L.Neidig,
and
E.I.Solomon
(2005).
Structure-function correlations in oxygen activating non-heme iron enzymes.
|
| |
Chem Commun (Camb),
(),
5843-5863.
|
 |
|
|
|
|
 |
N.J.Kershaw,
M.E.Caines,
M.C.Sleeman,
and
C.J.Schofield
(2005).
The enzymology of clavam and carbapenem biosynthesis.
|
| |
Chem Commun (Camb),
(),
4251-4263.
|
 |
|
|
|
|
 |
X.B.Wu,
K.Q.Fan,
Q.H.Wang,
and
K.Q.Yang
(2005).
C-terminus mutations of Acremonium chrysogenum deacetoxy/deacetylcephalosporin C synthase with improved activity toward penicillin analogs.
|
| |
FEMS Microbiol Lett,
246,
103-110.
|
 |
|
|
|
|
 |
C.J.Schofield,
and
P.J.Ratcliffe
(2004).
Oxygen sensing by HIF hydroxylases.
|
| |
Nat Rev Mol Cell Biol,
5,
343-354.
|
 |
|
|
|
|
 |
H.S.Chin,
K.S.Goo,
and
T.S.Sim
(2004).
A complete library of amino acid alterations at N304 in Streptomyces clavuligerus deacetoxycephalosporin C synthase elucidates the basis for enhanced penicillin analogue conversion.
|
| |
Appl Environ Microbiol,
70,
607-609.
|
 |
|
|
|
|
 |
I.Castro-Rodriguez,
H.Nakai,
L.N.Zakharov,
A.L.Rheingold,
and
K.Meyer
(2004).
A linear, O-coordinated eta1-CO2 bound to uranium.
|
| |
Science,
305,
1757-1759.
|
 |
|
|
|
|
 |
J.S.Hsu,
Y.B.Yang,
C.H.Deng,
C.L.Wei,
S.H.Liaw,
and
Y.C.Tsai
(2004).
Family shuffling of expandase genes to enhance substrate specificity for penicillin G.
|
| |
Appl Environ Microbiol,
70,
6257-6263.
|
 |
|
|
|
|
 |
K.Valegård,
A.C.Terwisscha van Scheltinga,
A.Dubus,
G.Ranghino,
L.M.Oster,
J.Hajdu,
and
I.Andersson
(2004).
The structural basis of cephalosporin formation in a mononuclear ferrous enzyme.
|
| |
Nat Struct Mol Biol,
11,
95.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.D.Lloyd,
S.J.Lipscomb,
K.S.Hewitson,
C.M.Hensgens,
J.E.Baldwin,
and
C.J.Schofield
(2004).
Controlling the substrate selectivity of deacetoxycephalosporin/deacetylcephalosporin C synthase.
|
| |
J Biol Chem,
279,
15420-15426.
|
 |
|
|
|
|
 |
C.L.Wei,
Y.B.Yang,
W.C.Wang,
W.C.Liu,
J.S.Hsu,
and
Y.C.Tsai
(2003).
Engineering Streptomyces clavuligerus deacetoxycephalosporin C synthase for optimal ring expansion activity toward penicillin G.
|
| |
Appl Environ Microbiol,
69,
2306-2312.
|
 |
|
|
|
|
 |
H.J.Lee,
Y.F.Dai,
C.Y.Shiau,
C.J.Schofield,
and
M.D.Lloyd
(2003).
The kinetic properties of various R258 mutants of deacetoxycephalosporin C synthase.
|
| |
Eur J Biochem,
270,
1301-1307.
|
 |
|
|
|
|
 |
M.J.Ryle,
K.D.Koehntop,
A.Liu,
L.Que,
and
R.P.Hausinger
(2003).
Interconversion of two oxidized forms of taurine/alpha-ketoglutarate dioxygenase, a non-heme iron hydroxylase: evidence for bicarbonate binding.
|
| |
Proc Natl Acad Sci U S A,
100,
3790-3795.
|
 |
|
|
|
|
 |
M.Mukherji,
C.J.Schofield,
A.S.Wierzbicki,
G.A.Jansen,
R.J.Wanders,
and
M.D.Lloyd
(2003).
The chemical biology of branched-chain lipid metabolism.
|
| |
Prog Lipid Res,
42,
359-376.
|
 |
|
|
|
|
 |
M.J.Ryle,
and
R.P.Hausinger
(2002).
Non-heme iron oxygenases.
|
| |
Curr Opin Chem Biol,
6,
193-201.
|
 |
|
|
|
|
 |
S.J.Lipscomb,
H.J.Lee,
M.Mukherji,
J.E.Baldwin,
C.J.Schofield,
and
M.D.Lloyd
(2002).
The role of arginine residues in substrate binding and catalysis by deacetoxycephalosporin C synthase.
|
| |
Eur J Biochem,
269,
2735-2739.
|
 |
|
|
|
|
 |
T.Duncan,
S.C.Trewick,
P.Koivisto,
P.A.Bates,
T.Lindahl,
and
B.Sedgwick
(2002).
Reversal of DNA alkylation damage by two human dioxygenases.
|
| |
Proc Natl Acad Sci U S A,
99,
16660-16665.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

