 |
PDBsum entry 1uo9

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
1uo9
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.14.20.1
- deacetoxycephalosporin-C synthase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Penicillin N and Deacetoxycephalosporin C Biosynthesis
|
 |
 |
 |
 |
 |
Reaction:
|
 |
penicillin N + 2-oxoglutarate + O2 = deacetoxycephalosporin C + succinate + CO2 + H2O
|
 |
 |
 |
 |
 |
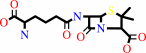 penicillin N
penicillin N
|
+
|
 2-oxoglutarate
2-oxoglutarate
|
+
|
 O2
O2
|
=
|
 deacetoxycephalosporin C
deacetoxycephalosporin C
|
+
|
 succinate
succinate
|
+
|
 CO2
CO2
|
+
|
H2O
Bound ligand (Het Group name = )
corresponds exactly
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Nat Struct Mol Biol
11:95
(2004)
|
|
PubMed id:
|
|
|
|

|
| |
|
The structural basis of cephalosporin formation in a mononuclear ferrous enzyme.
|
|
K.Valegård,
A.C.Terwisscha van Scheltinga,
A.Dubus,
G.Ranghino,
L.M.Oster,
J.Hajdu,
I.Andersson.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Deacetoxycephalosporin-C synthase (DAOCS) is a mononuclear ferrous enzyme that
transforms penicillins into cephalosporins by inserting a carbon atom into the
penicillin nucleus. In the first half-reaction, dioxygen and 2-oxoglutarate
produce a reactive iron-oxygen species, succinate and CO2. The oxidizing iron
species subsequently reacts with penicillin to give cephalosporin and water.
Here we describe high-resolution structures for ferrous DAOCS in complex with
penicillins, the cephalosporin product, the cosubstrate and the coproduct.
Steady-state kinetic data, quantum-chemical calculations and the new structures
indicate a reaction sequence in which a 'booby-trapped' oxidizing species is
formed. This species is stabilized by the negative charge of succinate on the
iron. The binding sites of succinate and penicillin overlap, and when penicillin
replaces succinate, it removes the stabilizing charge, eliciting oxidative
attack on itself. Requisite groups of penicillin are within 1 A of the expected
position of a ferryl oxygen in the enzyme-penicillin complex.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
Figure 1. The active site region of DAOCS in complex with
substrates and products. (a) The DAOCS -Fe(II)
-2-oxoglutarate complex2 at 1.5-Å resolution. (b) The DAOCS
-Fe(II) -succinate complex at 1.5-Å resolution. (c) The DAOCS
-Fe(II) -penicillin G complex at 1.6-Å resolution. (d) The DAOCS
-Fe(II) -2-oxoglutarate -penicillin G complex at 1.7 Å
resolution. (e) The DAOCS -Fe(II) -2-oxoglutarate -ampicillin
complex at 1.5-Å resolution. (f) The DAOCS -Fe(II) -DAOC complex
at 1.7-Å resolution. See text for details. The density next to
the penicillin side chain in d,e corresponds to a minor
alternative conformation of the side chain. Dioxygen is expected
to bind at the position of Wat1 in a. The oxygen of the ferryl
iron would be formed at this site^2. The carbon atoms in
2-oxoglutarate are yellow, in succinate orange, in penicillin G
magenta, in ampicillin cyan and in DAOC gold.
|
 |
Figure 5.
Figure 5. A possible mechanism for the ring expansion catalyzed
by DAOCS. The mechanism is based on the mode of penicillin
and cephalosporin binding shown in Figures 1d -f and 2. The
presumed oxidation states of the iron are marked. In the
oxidative half reaction, one of the oxygen atoms of dioxygen is
incorporated into succinate while the other one remains on the
iron. This oxygen can remove two electrons and two protons from
the five-membered thiazolidine ring to form the six-membered
dihydrothiazine ring of the cephalosporin product in the
reductive half reaction. Note ligation of the penicillin sulfur
to the iron (Fe -S distance: 2.1 -2.0 Å in the various
complexes) and that both  -
and -
and  -methyl
groups are in van der Waals contact with the iron (iron -methyl
distances: 2.1 -2.5 Å). -methyl
groups are in van der Waals contact with the iron (iron -methyl
distances: 2.1 -2.5 Å).
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from Macmillan Publishers Ltd:
Nat Struct Mol Biol
(2004,
11,
95-0)
copyright 2004.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
X.B.Wu,
X.Y.Tian,
J.J.Ji,
W.B.Wu,
K.Q.Fan,
and
K.Q.Yang
(2011).
Saturation mutagenesis of Acremonium chrysogenum deacetoxy/deacetylcephalosporin C synthase R308 site confirms its role in controlling substrate specificity.
|
| |
Biotechnol Lett,
33,
805-812.
|
 |
|
|
|
|
 |
P.He,
and
G.R.Moran
(2009).
We two alone will sing: the two-substrate alpha-keto acid-dependent oxygenases.
|
| |
Curr Opin Chem Biol,
13,
443-450.
|
 |
|
|
|
|
 |
K.S.Goo,
C.S.Chua,
and
T.S.Sim
(2008).
Relevant double mutations in bioengineered Streptomyces clavuligerus deacetoxycephalosporin C synthase result in higher binding specificities which improve penicillin bioconversion.
|
| |
Appl Environ Microbiol,
74,
1167-1175.
|
 |
|
|
|
|
 |
K.Sim Goo,
C.Song Chua,
and
T.S.Sim
(2008).
A complete library of amino acid alterations at R306 in Streptomyces clavuligerus deacetoxycephalosporin C synthase demonstrates its structural role in the ring-expansion activity.
|
| |
Proteins,
70,
739-747.
|
 |
|
|
|
|
 |
P.C.Bruijnincx,
G.van Koten,
and
R.J.Klein Gebbink
(2008).
Mononuclear non-heme iron enzymes with the 2-His-1-carboxylate facial triad: recent developments in enzymology and modeling studies.
|
| |
Chem Soc Rev,
37,
2716-2744.
|
 |
|
|
|
|
 |
K.S.Hewitson,
B.M.Liénard,
M.A.McDonough,
I.J.Clifton,
D.Butler,
A.S.Soares,
N.J.Oldham,
L.A.McNeill,
and
C.J.Schofield
(2007).
Structural and mechanistic studies on the inhibition of the hypoxia-inducible transcription factor hydroxylases by tricarboxylic acid cycle intermediates.
|
| |
J Biol Chem,
282,
3293-3301.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
V.Purpero,
and
G.R.Moran
(2007).
The diverse and pervasive chemistries of the alpha-keto acid dependent enzymes.
|
| |
J Biol Inorg Chem,
12,
587-601.
|
 |
|
|
|
|
 |
J.Ringvoll,
L.M.Nordstrand,
C.B.Vågbø,
V.Talstad,
K.Reite,
P.A.Aas,
K.H.Lauritzen,
N.B.Liabakk,
A.Bjørk,
R.W.Doughty,
P.Ã.˜.Falnes,
H.E.Krokan,
and
A.Klungland
(2006).
Repair deficient mice reveal mABH2 as the primary oxidative demethylase for repairing 1meA and 3meC lesions in DNA.
|
| |
EMBO J,
25,
2189-2198.
|
 |
|
|
|
|
 |
C.L.Wei,
Y.B.Yang,
C.H.Deng,
W.C.Liu,
J.S.Hsu,
Y.C.Lin,
S.H.Liaw,
and
Y.C.Tsai
(2005).
Directed evolution of Streptomyces clavuligerus deacetoxycephalosporin C synthase for enhancement of penicillin G expansion.
|
| |
Appl Environ Microbiol,
71,
8873-8880.
|
 |
|
|
|
|
 |
K.D.Koehntop,
J.P.Emerson,
and
L.Que
(2005).
The 2-His-1-carboxylate facial triad: a versatile platform for dioxygen activation by mononuclear non-heme iron(II) enzymes.
|
| |
J Biol Inorg Chem,
10,
87-93.
|
 |
|
|
|
|
 |
X.B.Wu,
K.Q.Fan,
Q.H.Wang,
and
K.Q.Yang
(2005).
C-terminus mutations of Acremonium chrysogenum deacetoxy/deacetylcephalosporin C synthase with improved activity toward penicillin analogs.
|
| |
FEMS Microbiol Lett,
246,
103-110.
|
 |
|
|
|
|
 |
J.S.Hsu,
Y.B.Yang,
C.H.Deng,
C.L.Wei,
S.H.Liaw,
and
Y.C.Tsai
(2004).
Family shuffling of expandase genes to enhance substrate specificity for penicillin G.
|
| |
Appl Environ Microbiol,
70,
6257-6263.
|
 |
|
|
|
|
 |
Z.Zhang,
J.S.Ren,
I.J.Clifton,
and
C.J.Schofield
(2004).
Crystal structure and mechanistic implications of 1-aminocyclopropane-1-carboxylic acid oxidase--the ethylene-forming enzyme.
|
| |
Chem Biol,
11,
1383-1394.
|
 |
|
PDB codes:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

