 |
PDBsum entry 1dd2

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.1.3.1
- methylmalonyl-CoA carboxytransferase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
(S)-methylmalonyl-CoA + pyruvate = propanoyl-CoA + oxaloacetate
|
 |
 |
 |
 |
 |
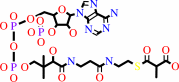 (S)-methylmalonyl-CoA
(S)-methylmalonyl-CoA
|
+
|
 pyruvate
pyruvate
|
=
|
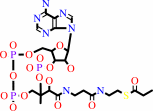 propanoyl-CoA
propanoyl-CoA
|
+
|
 oxaloacetate
oxaloacetate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Biotin; Cobalt cation; Zn(2+)
|
 |
 |
 |
 |
 |
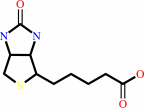 Biotin
Biotin
|
Cobalt cation
|
Zn(2+)
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Biochemistry
39:2509-2516
(2000)
|
|
PubMed id:
|
|
|
|

|
| |
|
High resolution solution structure of the 1.3S subunit of transcarboxylase from Propionibacterium shermanii.
|
|
D.V.Reddy,
B.C.Shenoy,
P.R.Carey,
F.D.Sönnichsen.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Transcarboxylase (TC) from Propionibacterium shermanii, a biotin-dependent
enzyme, catalyzes the transfer of a carboxyl group from methylmalonyl-CoA to
pyruvate to form propionyl-CoA and oxalacetate. Within the multi-subunit enzyme
complex, the 1.3S subunit functions as the carboxyl group carrier and also binds
the other two subunits to assist in the overall assembly of the enzyme. The 1.3S
subunit is a 123 amino acid polypeptide (12.6 kDa) to which biotin is covalently
attached at Lys 89. The three-dimensional solution structure of the full-length
holo-1.3S subunit of TC has been solved by multidimensional heteronuclear NMR
spectroscopy. The C-terminal half of the protein (51-123) is folded into a
compact all-beta-domain comprising of two four-stranded antiparallel beta-sheets
connected by short loops and turns. The fold exhibits a high 2-fold internal
symmetry and is similar to that of the biotin carboxyl carrier protein (BCCP) of
acetyl-CoA carboxylase, but lacks an extension that has been termed "protruding
thumb" in BCCP. The first 50 residues, which have been shown to be involved in
intersubunit interactions in the intact enzyme, appear to be disordered in the
isolated 1.3S subunit. The molecular surface of the folded domain has two
distinct surfaces: one side is highly charged, while the other comprises mainly
hydrophobic, highly conserved residues.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
G.Gago,
L.Diacovich,
A.Arabolaza,
S.C.Tsai,
and
H.Gramajo
(2011).
Fatty acid biosynthesis in actinomycetes.
|
| |
FEMS Microbiol Rev,
35,
475-497.
|
 |
|
|
|
|
 |
J.E.Butler,
N.D.Young,
and
D.R.Lovley
(2009).
Evolution from a respiratory ancestor to fill syntrophic and fermentative niches: comparative fenomics of six Geobacteraceae species.
|
| |
BMC Genomics,
10,
103.
|
 |
|
|
|
|
 |
B.Bagautdinov,
Y.Matsuura,
S.Bagautdinova,
and
N.Kunishima
(2008).
Protein biotinylation visualized by a complex structure of biotin protein ligase with a substrate.
|
| |
J Biol Chem,
283,
14739-14750.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
C.K.Lee,
H.K.Cheong,
K.S.Ryu,
J.I.Lee,
W.Lee,
Y.H.Jeon,
and
C.Cheong
(2008).
Biotinoyl domain of human acetyl-CoA carboxylase: Structural insights into the carboxyl transfer mechanism.
|
| |
Proteins,
72,
613-624.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
I.Chen,
Y.A.Choi,
and
A.Y.Ting
(2007).
Phage display evolution of a peptide substrate for yeast biotin ligase and application to two-color quantum dot labeling of cell surface proteins.
|
| |
J Am Chem Soc,
129,
6619-6625.
|
 |
|
|
|
|
 |
N.M.Lorenzon,
and
K.G.Beam
(2007).
Accessibility of targeted DHPR sites to streptavidin and functional effects of binding on EC coupling.
|
| |
J Gen Physiol,
130,
379-388.
|
 |
|
|
|
|
 |
S.K.Campos,
and
M.A.Barry
(2007).
Current advances and future challenges in Adenoviral vector biology and targeting.
|
| |
Curr Gene Ther,
7,
189-204.
|
 |
|
|
|
|
 |
E.D.Streaker,
and
D.Beckett
(2006).
Nonenzymatic biotinylation of a biotin carboxyl carrier protein: unusual reactivity of the physiological target lysine.
|
| |
Protein Sci,
15,
1928-1935.
|
 |
|
|
|
|
 |
G.Cui,
B.Nan,
J.Hu,
Y.Wang,
C.Jin,
and
B.Xia
(2006).
Identification and solution structures of a single domain biotin/lipoyl attachment protein from Bacillus subtilis.
|
| |
J Biol Chem,
281,
20598-20607.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
Y.Maegawa,
H.Morita,
D.Iyaguchi,
M.Yao,
N.Watanabe,
and
I.Tanaka
(2006).
Structure of the catalytic nucleotide-binding subunit A of A-type ATP synthase from Pyrococcus horikoshii reveals a novel domain related to the peripheral stalk.
|
| |
Acta Crystallogr D Biol Crystallogr,
62,
483-488.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
E.Choi-Rhee,
H.Schulman,
and
J.E.Cronan
(2004).
Promiscuous protein biotinylation by Escherichia coli biotin protein ligase.
|
| |
Protein Sci,
13,
3043-3050.
|
 |
|
|
|
|
 |
N.M.Lorenzon,
C.S.Haarmann,
E.E.Norris,
S.Papadopoulos,
and
K.G.Beam
(2004).
Metabolic biotinylation as a probe of supramolecular structure of the triad junction in skeletal muscle.
|
| |
J Biol Chem,
279,
44057-44064.
|
 |
|
|
|
|
 |
P.R.Hall,
R.Zheng,
M.Pusztai-Carey,
F.van den Akker,
P.R.Carey,
and
V.C.Yee
(2004).
Expression and crystallization of several forms of the Propionibacterium shermanii transcarboxylase 5S subunit.
|
| |
Acta Crystallogr D Biol Crystallogr,
60,
521-523.
|
 |
|
|
|
|
 |
S.K.Campos,
and
M.A.Barry
(2004).
Rapid construction of capsid-modified adenoviral vectors through bacteriophage lambda Red recombination.
|
| |
Hum Gene Ther,
15,
1125-1130.
|
 |
|
|
|
|
 |
K.S.Wendt,
I.Schall,
R.Huber,
W.Buckel,
and
U.Jacob
(2003).
Crystal structure of the carboxyltransferase subunit of the bacterial sodium ion pump glutaconyl-coenzyme A decarboxylase.
|
| |
EMBO J,
22,
3493-3502.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
P.R.Hall,
Y.F.Wang,
R.E.Rivera-Hainaj,
X.Zheng,
M.Pustai-Carey,
P.R.Carey,
and
V.C.Yee
(2003).
Transcarboxylase 12S crystal structure: hexamer assembly and substrate binding to a multienzyme core.
|
| |
EMBO J,
22,
2334-2347.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
J.E.Cronan
(2002).
Interchangeable enzyme modules. Functional replacement of the essential linker of the biotinylated subunit of acetyl-CoA carboxylase with a linker from the lipoylated subunit of pyruvate dehydrogenase.
|
| |
J Biol Chem,
277,
22520-22527.
|
 |
|
|
|
|
 |
J.E.Cronan,
and
G.L.Waldrop
(2002).
Multi-subunit acetyl-CoA carboxylases.
|
| |
Prog Lipid Res,
41,
407-435.
|
 |
|
|
|
|
 |
Y.F.Wang,
D.C.Hyatt,
R.E.Rivera,
P.R.Carey,
and
V.C.Yee
(2001).
Crystallization and preliminary X-ray analysis of the 12S central subunit of transcarboxylase from Propionibacterium shermanii.
|
| |
Acta Crystallogr D Biol Crystallogr,
57,
266-268.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

