 |
PDBsum entry 2kcc

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Ligase
|
 |
|
Title:
|
 |
Solution structure of biotinoyl domain from human acetyl-coa carboxylase 2
|
|
Structure:
|
 |
Acetyl-coa carboxylase 2. Chain: a. Fragment: biotinoyl domain, residues 891-964. Synonym: acc-beta, biotin carboxylase. Engineered: yes
|
|
Source:
|
 |
Homo sapiens. Human. Organism_taxid: 9606. Gene: acacb, acc2, accb. Expressed in: escherichia coli. Expression_system_taxid: 562.
|
|
NMR struc:
|
 |
20 models
|
 |
|
Authors:
|
 |
C.Lee,H.Cheong,K.Ryu,J.Lee,W.Lee,Y.Jeon,C.Cheong
|
Key ref:

|
 |
C.K.Lee
et al.
(2008).
Biotinoyl domain of human acetyl-CoA carboxylase: Structural insights into the carboxyl transfer mechanism.
Proteins,
72,
613-624.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
19-Dec-08
|
Release date:
|
17-Feb-09
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
O00763
(ACACB_HUMAN) -
Acetyl-CoA carboxylase 2 from Homo sapiens
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
2458 a.a.
84 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|
*
PDB and UniProt seqs differ
at 8 residue positions (black
crosses)
|
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.6.4.1.2
- acetyl-CoA carboxylase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
hydrogencarbonate + acetyl-CoA + ATP = malonyl-CoA + ADP + phosphate + H+
|
 |
 |
 |
 |
 |
 hydrogencarbonate
hydrogencarbonate
|
+
|
 acetyl-CoA
acetyl-CoA
|
+
|
 ATP
ATP
|
=
|
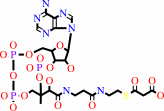 malonyl-CoA
malonyl-CoA
|
+
|
 ADP
ADP
|
+
|
 phosphate
phosphate
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Biotin
|
 |
 |
 |
 |
 |
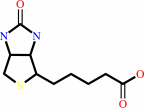 Biotin
Biotin
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Proteins
72:613-624
(2008)
|
|
PubMed id:
|
|
|
|

|
| |
|
Biotinoyl domain of human acetyl-CoA carboxylase: Structural insights into the carboxyl transfer mechanism.
|
|
C.K.Lee,
H.K.Cheong,
K.S.Ryu,
J.I.Lee,
W.Lee,
Y.H.Jeon,
C.Cheong.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Acetyl-CoA carboxylase (ACC) catalyzes the first step in fatty acid
biosynthesis: the synthesis of malonyl-CoA from acetyl-CoA. As essential
regulators of fatty acid biosynthesis and metabolism, ACCs are regarded as
therapeutic targets for the treatment of metabolic diseases such as obesity. In
ACC, the biotinoyl domain performs a critical function by transferring an
activated carboxyl group from the biotin carboxylase domain to the carboxyl
transferase domain, followed by carboxyl transfer to malonyl-CoA. Despite the
intensive research on this enzyme, only the bacterial and yeast ACC structures
are currently available. To explore the mechanism of ACC holoenzyme function, we
determined the structure of the biotinoyl domain of human ACC2 and analyzed its
characteristics and interaction with the biotin ligase, BirA using NMR
spectroscopy. The 3D structure of the hACC2 biotinoyl domain has a similar
folding topology to the earlier determined domains from E. coli and P.
shermanii. However, the local structures near the biotinylation sites have
notable differences that include the geometry of the consensus "Met-Lys-Met"
(MKM) motif and the absence of "thumb" structure in the hACC2 biotinoyl domain.
Observations of the NMR signals upon the biotinylation indicate that the biotin
group of hACC2 does not affect the structure of the biotinoyl domain, while the
biotin group for E. coli ACC interacts directly with the thumb residues that are
not present in the hACC2 structure. These results imply that, in the E. coli ACC
reaction, the biotin moiety carrying the carboxyl group from BC to CT can pause
at the thumb of the BCCP domain. The human biotinoyl domain, however, lacks the
thumb structure and does not have additional noncovalent interactions with the
biotin moiety; thus, the flexible motion of the biotinylated lysine residue must
underlie the "swinging arm" motion. The chemical shift perturbation and the
cross saturation experiments of the human ACC2 holo-biotinoyl upon the addition
of the biotin ligase (BirA) showed the interaction surface near the MKM motif,
the two glutamic acids (Glu 926, Glu 953), and the positively charged residues
(several lysine and arginine residues). This study provides insight into the
mechanism of ACC holoenzyme function and supports the swinging arm model in
human ACCs.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 2.
Figure 2. The 20 lowest-energy solution structure of the human
ACC2 biotinoyl domain (residues 891-964). A: Twenty structures
of hACC2 biotinoyl domain are superimposed (residue range for
r.m.s.d. calculation and matching is 895-960). The backbone
region was well converged, and the r.m.s.d. was 0.23 Å. B:
Ribbon representation of the hACC2 biotinoyl domain. The two
 -sheets
are orange and green, respectively. C: Side chains of
hydrophobic residues involved in forming the hydrophobic core.
D: Five bifurcated hydrogen bonds (dotted lines) stabilize the -sheets
are orange and green, respectively. C: Side chains of
hydrophobic residues involved in forming the hydrophobic core.
D: Five bifurcated hydrogen bonds (dotted lines) stabilize the
 -sandwich
structure. -sandwich
structure.
|
 |
Figure 4.
Figure 4. The surface representation map of the biotinoyl
domain from three different species. The surface representation
map of the biotinoyl domain from three different species is
shown with the biotin protein ligase recognition motif,
Met-Lys-Met. Red and blue represent negatively and positively
charged residues, respectively.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from John Wiley & Sons, Inc.:
Proteins
(2008,
72,
613-624)
copyright 2008.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

