 |
PDBsum entry 1add

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Hydrolase(acting in cyclicamidines)
|
PDB id
|
|

|

|
1add
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.3.5.4.4
- adenosine deaminase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
|
1.
|
adenosine + H2O + H+ = inosine + NH4+
|
|
2.
|
2'-deoxyadenosine + H2O + H+ = 2'-deoxyinosine + NH4+
|
|
 |
 |
 |
 |
 |
adenosine
Bound ligand (Het Group name = )
matches with 90.00% similarity
|
+
|
H2O
|
+
|
H(+)
|
=
|
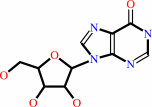 inosine
inosine
|
+
|
NH4(+)
|
|
 |
 |
 |
 |
 |
2'-deoxyadenosine
Bound ligand (Het Group name = )
matches with 85.00% similarity
|
+
|
H2O
|
+
|
H(+)
|
=
|
2'-deoxyinosine
|
+
|
NH4(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Biochemistry
32:1689-1694
(1993)
|
|
PubMed id:
|
|
|
|

|
| |
|
A pre-transition-state mimic of an enzyme: X-ray structure of adenosine deaminase with bound 1-deazaadenosine and zinc-activated water.
|
|
D.K.Wilson,
F.A.Quiocho.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The refined 2.4-A structure of adenosine deaminase, recently discovered to be a
zinc metalloenzyme [Wilson, D. K., Rudolph, F. B., & Quiocho, F. A. (1991)
Science 252, 1278-1284], complexed with the ground-state analog 1-deazaadenosine
shows the mode of binding of the analog and, unexpectedly, a zinc-activated
water (hydroxide). This structure of a pre-transition-state mimic, combined with
that previously determined for the complex with 6(R)-hydroxy-1,6-dihydropurine
ribonucleoside, a nearly ideal transition-state analog, sheds new understanding
of the precise stereospecificity and hydrolytic catalysis of an important and
well-characterized member of a large group of zinc metalloenzymes. As both of
these excellent mimics were generated in the active site, they demonstrate a
powerful means of dissecting the course of an enzymatic reaction by direct
crystallographic analysis.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
M.A.Zoroddu,
S.Medici,
M.Peana,
and
R.Anedda
(2010).
NMR studies of zinc binding in a multi-histidinic peptide fragment.
|
| |
Dalton Trans,
39,
1282-1294.
|
 |
|
|
|
|
 |
P.Bhattacharjee,
and
R.Sharma
(2009).
Antithetical effects of corticosterone and dibutyryl cAMP on adenosine deaminase in the gastrointestinal tract of chicken during postnatal development.
|
| |
Mol Cell Biochem,
327,
79-86.
|
 |
|
|
|
|
 |
A.D.Hill,
and
P.J.Reilly
(2008).
A Gibbs free energy correlation for automated docking of carbohydrates.
|
| |
J Comput Chem,
29,
1131-1141.
|
 |
|
|
|
|
 |
E.T.Larson,
W.Deng,
B.E.Krumm,
A.Napuli,
N.Mueller,
W.C.Van Voorhis,
F.S.Buckner,
E.Fan,
A.Lauricella,
G.DeTitta,
J.Luft,
F.Zucker,
W.G.Hol,
C.L.Verlinde,
and
E.A.Merritt
(2008).
Structures of substrate- and inhibitor-bound adenosine deaminase from a human malaria parasite show a dramatic conformational change and shed light on drug selectivity.
|
| |
J Mol Biol,
381,
975-988.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
J.L.Seffernick,
A.Aleem,
J.P.Osborne,
G.Johnson,
M.J.Sadowsky,
and
L.P.Wackett
(2007).
Hydroxyatrazine N-ethylaminohydrolase (AtzB): an amidohydrolase superfamily enzyme catalyzing deamination and dechlorination.
|
| |
J Bacteriol,
189,
6989-6997.
|
 |
|
|
|
|
 |
M.Luo,
V.Singh,
E.A.Taylor,
and
V.L.Schramm
(2007).
Transition-state variation in human, bovine, and Plasmodium falciparum adenosine deaminases.
|
| |
J Am Chem Soc,
129,
8008-8017.
|
 |
|
|
|
|
 |
I.A.Il'icheva,
I.u.P.Zarubin,
P.A.Kostin,
D.V.Mirgorodskiĭ,
P.P.Purygin,
and
V.L.Florent'ev
(2005).
[Theoretical study of the structure of adenosine deaminase complexes with adenosine analogues: I. Aza-, deaza- and isomeric azadeazaanalogues of adenosine]
|
| |
Bioorg Khim,
31,
488-502.
|
 |
|
|
|
|
 |
A.A.Saboury,
S.Bagheri,
G.Ataie,
M.Amanlou,
A.A.Moosavi-Movahedi,
G.H.Hakimelahi,
G.Cristalli,
and
S.Namaki
(2004).
Binding properties of adenosine deaminase interacted with theophylline.
|
| |
Chem Pharm Bull (Tokyo),
52,
1179-1182.
|
 |
|
|
|
|
 |
H.Li,
H.Xu,
D.E.Graham,
and
R.H.White
(2003).
The Methanococcus jannaschii dCTP deaminase is a bifunctional deaminase and diphosphatase.
|
| |
J Biol Chem,
278,
11100-11106.
|
 |
|
|
|
|
 |
S.B.Mulrooney,
and
R.P.Hausinger
(2003).
Metal ion dependence of recombinant Escherichia coli allantoinase.
|
| |
J Bacteriol,
185,
126-134.
|
 |
|
|
|
|
 |
T.Kinoshita,
N.Nishio,
I.Nakanishi,
A.Sato,
and
T.Fujii
(2003).
Structure of bovine adenosine deaminase complexed with 6-hydroxy-1,6-dihydropurine riboside.
|
| |
Acta Crystallogr D Biol Crystallogr,
59,
299-303.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
N.Shapir,
J.P.Osborne,
G.Johnson,
M.J.Sadowsky,
and
L.P.Wackett
(2002).
Purification, substrate range, and metal center of AtzC: the N-isopropylammelide aminohydrolase involved in bacterial atrazine metabolism.
|
| |
J Bacteriol,
184,
5376-5384.
|
 |
|
|
|
|
 |
G.Cristalli,
S.Costanzi,
C.Lambertucci,
G.Lupidi,
S.Vittori,
R.Volpini,
and
E.Camaioni
(2001).
Adenosine deaminase: functional implications and different classes of inhibitors.
|
| |
Med Res Rev,
21,
105-128.
|
 |
|
|
|
|
 |
J.A.Gerlt,
and
P.C.Babbitt
(2001).
Divergent evolution of enzymatic function: mechanistically diverse superfamilies and functionally distinct suprafamilies.
|
| |
Annu Rev Biochem,
70,
209-246.
|
 |
|
|
|
|
 |
J.L.Seffernick,
M.L.de Souza,
M.J.Sadowsky,
and
L.P.Wackett
(2001).
Melamine deaminase and atrazine chlorohydrolase: 98 percent identical but functionally different.
|
| |
J Bacteriol,
183,
2405-2410.
|
 |
|
|
|
|
 |
H.Ford,
F.Dai,
L.Mu,
M.A.Siddiqui,
M.C.Nicklaus,
L.Anderson,
V.E.Marquez,
and
J.J.Barchi
(2000).
Adenosine deaminase prefers a distinct sugar ring conformation for binding and catalysis: kinetic and structural studies.
|
| |
Biochemistry,
39,
2581-2592.
|
 |
|
|
|
|
 |
T.Kinoshita,
N.Nishio,
A.Sato,
and
M.Murata
(1999).
Crystallization and preliminary analysis of bovine adenosine deaminase.
|
| |
Acta Crystallogr D Biol Crystallogr,
55,
2031-2032.
|
 |
|
|
|
|
 |
H.Deng,
L.C.Kurz,
F.B.Rudolph,
and
R.Callender
(1998).
Characterization of hydrogen bonding in the complex of adenosine deaminase with a transition state analogue: a Raman spectroscopic study.
|
| |
Biochemistry,
37,
4968-4976.
|
 |
|
|
|
|
 |
J.A.Gerlt,
and
P.C.Babbitt
(1998).
Mechanistically diverse enzyme superfamilies: the importance of chemistry in the evolution of catalysis.
|
| |
Curr Opin Chem Biol,
2,
607-612.
|
 |
|
|
|
|
 |
L.J.Jiang,
W.Maret,
and
B.L.Vallee
(1998).
The ATP-metallothionein complex.
|
| |
Proc Natl Acad Sci U S A,
95,
9146-9149.
|
 |
|
|
|
|
 |
R.Franco,
A.Valenzuela,
C.Lluis,
and
J.Blanco
(1998).
Enzymatic and extraenzymatic role of ecto-adenosine deaminase in lymphocytes.
|
| |
Immunol Rev,
161,
27-42.
|
 |
|
|
|
|
 |
Z.Wang,
and
F.A.Quiocho
(1998).
Complexes of adenosine deaminase with two potent inhibitors: X-ray structures in four independent molecules at pH of maximum activity.
|
| |
Biochemistry,
37,
8314-8324.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
B.F.Cooper,
V.Sideraki,
D.K.Wilson,
D.Y.Dominguez,
S.W.Clark,
F.A.Quiocho,
and
F.B.Rudolph
(1997).
The role of divalent cations in structure and function of murine adenosine deaminase.
|
| |
Protein Sci,
6,
1031-1037.
|
 |
|
|
|
|
 |
R.Franco,
V.Casadó,
F.Ciruela,
C.Saura,
J.Mallol,
E.I.Canela,
and
C.Lluis
(1997).
Cell surface adenosine deaminase: much more than an ectoenzyme.
|
| |
Prog Neurobiol,
52,
283-294.
|
 |
|
|
|
|
 |
S.Xiang,
S.A.Short,
R.Wolfenden,
and
C.W.Carter
(1997).
The structure of the cytidine deaminase-product complex provides evidence for efficient proton transfer and ground-state destabilization.
|
| |
Biochemistry,
36,
4768-4774.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
X.Zhao,
J.Liu,
D.S.Hsu,
S.Zhao,
J.S.Taylor,
and
A.Sancar
(1997).
Reaction mechanism of (6-4) photolyase.
|
| |
J Biol Chem,
272,
32580-32590.
|
 |
|
|
|
|
 |
E.Jabri,
and
P.A.Karplus
(1996).
Structures of the Klebsiella aerogenes urease apoenzyme and two active-site mutants.
|
| |
Biochemistry,
35,
10616-10626.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
H.Grosjean,
S.Auxilien,
F.Constantinesco,
C.Simon,
Y.Corda,
H.F.Becker,
D.Foiret,
A.Morin,
Y.X.Jin,
M.Fournier,
and
J.L.Fourrey
(1996).
Enzymatic conversion of adenosine to inosine and to N1-methylinosine in transfer RNAs: a review.
|
| |
Biochimie,
78,
488-501.
|
 |
|
|
|
|
 |
K.A.Mohamedali,
L.C.Kurz,
and
F.B.Rudolph
(1996).
Site-directed mutagenesis of active site glutamate-217 in mouse adenosine deaminase.
|
| |
Biochemistry,
35,
1672-1680.
|
 |
|
|
|
|
 |
R.A.Laskowski,
N.M.Luscombe,
M.B.Swindells,
and
J.M.Thornton
(1996).
Protein clefts in molecular recognition and function.
|
| |
Protein Sci,
5,
2438-2452.
|
 |
|
|
|
|
 |
S.Xiang,
S.A.Short,
R.Wolfenden,
and
C.W.Carter
(1996).
Cytidine deaminase complexed to 3-deazacytidine: a "valence buffer" in zinc enzyme catalysis.
|
| |
Biochemistry,
35,
1335-1341.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
V.Sideraki,
D.K.Wilson,
L.C.Kurz,
F.A.Quiocho,
and
F.B.Rudolph
(1996).
Site-directed mutagenesis of histidine 238 in mouse adenosine deaminase: substitution of histidine 238 does not impede hydroxylate formation.
|
| |
Biochemistry,
35,
15019-15028.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
V.Sideraki,
K.A.Mohamedali,
D.K.Wilson,
Z.Chang,
R.E.Kellems,
F.A.Quiocho,
and
F.B.Rudolph
(1996).
Probing the functional role of two conserved active site aspartates in mouse adenosine deaminase.
|
| |
Biochemistry,
35,
7862-7872.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
I.Santisteban,
F.X.Arredondo-Vega,
S.Kelly,
M.Debre,
A.Fischer,
J.L.Pérignon,
B.Hilman,
J.elDahr,
D.H.Dreyfus,
and
E.W.Gelfand
(1995).
Four new adenosine deaminase mutations, altering a zinc-binding histidine, two conserved alanines, and a 5' splice site.
|
| |
Hum Mutat,
5,
243-250.
|
 |
|
|
|
|
 |
L.S.Singh,
and
R.Sharma
(1995).
Developmental expression and corticosterone inhibition of adenosine deaminase activity in different tissues of mice.
|
| |
Mech Ageing Dev,
80,
85-92.
|
 |
|
|
|
|
 |
D.K.Wilson,
and
F.A.Quiocho
(1994).
Crystallographic observation of a trapped tetrahedral intermediate in a metalloenzyme.
|
| |
Nat Struct Biol,
1,
691-694.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

