 |
PDBsum entry 1af2

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.3.5.4.5
- cytidine deaminase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
|
1.
|
cytidine + H2O + H+ = uridine + NH4+
|
|
2.
|
2'-deoxycytidine + H2O + H+ = 2'-deoxyuridine + NH4+
|
|
 |
 |
 |
 |
 |
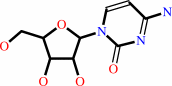 cytidine
cytidine
|
+
|
H2O
|
+
|
H(+)
|
=
|
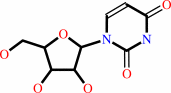 uridine
uridine
|
+
|
NH4(+)
Bound ligand (Het Group name = )
corresponds exactly
|
|
 |
 |
 |
 |
 |
 2'-deoxycytidine
2'-deoxycytidine
|
+
|
H2O
|
+
|
H(+)
|
=
|
2'-deoxyuridine
Bound ligand (Het Group name = )
matches with 94.12% similarity
|
+
|
NH4(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Zn(2+)
|
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Biochemistry
36:4768-4774
(1997)
|
|
PubMed id:
|
|
|
|

|
| |
|
The structure of the cytidine deaminase-product complex provides evidence for efficient proton transfer and ground-state destabilization.
|
|
S.Xiang,
S.A.Short,
R.Wolfenden,
C.W.Carter.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Crystal structures of the cytidine deaminase-uridine product complex prepared
either by cocrystallizing enzyme with uridine or by diffusing cytidine into
ligand-free crystals show that the product binds as a 4-ketopyrimidine. They
reveal four additional features of the catalytic process. (1) A water molecule
bound to a site previously observed to bind the incoming 4-NH2 group represents
the site for the leaving ammonia molecule. The conserved Pro 128 accommodates
both moieties by orienting the carbonyl group of the previous residue. (2) The
Glu 104 carboxylate group rotates from its hydrogen bond to the O4 hydroxyl
group in transition-state analog complexes, forming a new hydrogen bond to the
leaving group moiety. Thus, after stabilizing the hydroxyl group in the
transition state, Glu 104 transfers a proton from that group to the leaving
amino group, promoting enol-to-keto isomerization of the product. (3) Difference
Fourier comparisons with transition-state complexes indicate that the pyrimidine
ring rotates toward the zinc by approximately 10 degrees. The active site thus
"pulls" the ring and 4-NH2 group in opposite directions during catalysis. To
preserve coplanarity of the 4-keto group with the pyrimidine ring, the N1-C1'
glycosidic bond bends by approximately 19 degrees out of the ring plane. This
distortion may "spring-load" the product complex and promote dissociation.
Failure to recognize a similar distortion could explain an earlier
crystallographic interpretation of the adenosine deaminase-inosine complex
[Wilson, D. K., & Quiocho, F. A. (1994) Nat. Struct. Biol. 1, 691-694]. (4)
The Zn-Sgamma132 bond, which lengthens in transition-state complexes, shortens
as the O4 atom returns to a state of lower negative charge in the planar
product, consistent with our previous proposal that this bond buffers the zinc
bond valence, compensating buildup of negative charge on the oxygen nucleophile
during catalysis.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
X.Li,
S.A.Hayik,
and
K.M.Merz
(2010).
QM/MM X-ray refinement of zinc metalloenzymes.
|
| |
J Inorg Biochem,
104,
512-522.
|
 |
|
|
|
|
 |
A.Furukawa,
T.Nagata,
A.Matsugami,
Y.Habu,
R.Sugiyama,
F.Hayashi,
N.Kobayashi,
S.Yokoyama,
H.Takaku,
and
M.Katahira
(2009).
Structure, interaction and real-time monitoring of the enzymatic reaction of wild-type APOBEC3G.
|
| |
EMBO J,
28,
440-451.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
C.K.Lee,
H.K.Cheong,
K.S.Ryu,
J.I.Lee,
W.Lee,
Y.H.Jeon,
and
C.Cheong
(2008).
Biotinoyl domain of human acetyl-CoA carboxylase: Structural insights into the carboxyl transfer mechanism.
|
| |
Proteins,
72,
613-624.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
K.M.Chen,
E.Harjes,
P.J.Gross,
A.Fahmy,
Y.Lu,
K.Shindo,
R.S.Harris,
and
H.Matsuo
(2008).
Structure of the DNA deaminase domain of the HIV-1 restriction factor APOBEC3G.
|
| |
Nature,
452,
116-119.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
T.Kumasaka,
M.Yamamoto,
M.Furuichi,
M.Nakasako,
A.H.Teh,
M.Kimura,
I.Yamaguchi,
and
T.Ueki
(2007).
Crystal Structures of Blasticidin S Deaminase (BSD): IMPLICATIONS FOR DYNAMIC PROPERTIES OF CATALYTIC ZINC.
|
| |
J Biol Chem,
282,
37103-37111.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
C.H.Borchers,
V.E.Marquez,
G.K.Schroeder,
S.A.Short,
M.J.Snider,
J.P.Speir,
and
R.Wolfenden
(2004).
Fourier transform ion cyclotron resonance MS reveals the presence of a water molecule in an enzyme transition-state analogue complex.
|
| |
Proc Natl Acad Sci U S A,
101,
15341-15345.
|
 |
|
|
|
|
 |
S.H.Liaw,
Y.J.Chang,
C.T.Lai,
H.C.Chang,
and
G.G.Chang
(2004).
Crystal structure of Bacillus subtilis guanine deaminase: the first domain-swapped structure in the cytidine deaminase superfamily.
|
| |
J Biol Chem,
279,
35479-35485.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
G.C.Ireton,
M.E.Black,
and
B.L.Stoddard
(2003).
The 1.14 A crystal structure of yeast cytosine deaminase: evolution of nucleotide salvage enzymes and implications for genetic chemotherapy.
|
| |
Structure,
11,
961-972.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
H.Li,
H.Xu,
D.E.Graham,
and
R.H.White
(2003).
The Methanococcus jannaschii dCTP deaminase is a bifunctional deaminase and diphosphatase.
|
| |
J Biol Chem,
278,
11100-11106.
|
 |
|
|
|
|
 |
T.P.Ko,
J.J.Lin,
C.Y.Hu,
Y.H.Hsu,
A.H.Wang,
and
S.H.Liaw
(2003).
Crystal structure of yeast cytosine deaminase. Insights into enzyme mechanism and evolution.
|
| |
J Biol Chem,
278,
19111-19117.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
M.J.Snider,
D.Lazarevic,
and
R.Wolfenden
(2002).
Catalysis by entropic effects: the action of cytidine deaminase on 5,6-dihydrocytidine.
|
| |
Biochemistry,
41,
3925-3930.
|
 |
|
|
|
|
 |
R.C.Noonan,
C.W.Carter CW,
and
C.K.Bagdassarian
(2002).
Enzymatic conformational fluctuations along the reaction coordinate of cytidine deaminase.
|
| |
Protein Sci,
11,
1424-1434.
|
 |
|
|
|
|
 |
K.O.Alper,
M.Singla,
J.L.Stone,
and
C.K.Bagdassarian
(2001).
Correlated conformational fluctuations during enzymatic catalysis: Implications for catalytic rate enhancement.
|
| |
Protein Sci,
10,
1319-1330.
|
 |
|
|
|
|
 |
M.J.Snider,
S.Gaunitz,
C.Ridgway,
S.A.Short,
and
R.Wolfenden
(2000).
Temperature effects on the catalytic efficiency, rate enhancement, and transition state affinity of cytidine deaminase, and the thermodynamic consequences for catalysis of removing a substrate "anchor".
|
| |
Biochemistry,
39,
9746-9753.
|
 |
|
|
|
|
 |
J.C.Milne,
R.S.Roy,
A.C.Eliot,
N.L.Kelleher,
A.Wokhlu,
B.Nickels,
and
C.T.Walsh
(1999).
Cofactor requirements and reconstitution of microcin B17 synthetase: a multienzyme complex that catalyzes the formation of oxazoles and thiazoles in the antibiotic microcin B17.
|
| |
Biochemistry,
38,
4768-4781.
|
 |
|
|
|
|
 |
D.C.Carlow,
S.A.Short,
and
R.Wolfenden
(1998).
Complementary truncations of a hydrogen bond to ribose involved in transition-state stabilization by cytidine deaminase.
|
| |
Biochemistry,
37,
1199-1203.
|
 |
|
|
|
|
 |
D.Carlow,
and
R.Wolfenden
(1998).
Substrate connectivity effects in the transition state for cytidine deaminase.
|
| |
Biochemistry,
37,
11873-11878.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

