 |
PDBsum entry 2dvt

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.4.1.1.103
- gamma-resorcylate decarboxylase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
2,6-dihydroxybenzoate + H+ = resorcinol + CO2
|
 |
 |
 |
 |
 |
2,6-dihydroxybenzoate
|
+
|
H(+)
|
=
|
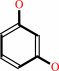 resorcinol
resorcinol
|
+
|
 CO2
CO2
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Zn(2+)
|
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Biol Chem
281:34365-34373
(2006)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structures of nonoxidative zinc-dependent 2,6-dihydroxybenzoate (gamma-resorcylate) decarboxylase from Rhizobium sp. strain MTP-10005.
|
|
M.Goto,
H.Hayashi,
I.Miyahara,
K.Hirotsu,
M.Yoshida,
T.Oikawa.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Reversible 2,6-dihydroxybenzoate decarboxylase from Rhizobium sp. strain
MTP-10005 belongs to a nonoxidative decarboxylase family. We have determined the
structures of the following three forms of the enzyme: the native form, the
complex with the true substrate (2,6-dihydroxybenzoate), and the complex with
2,3-dihydroxybenzaldehyde at 1.7-, 1.9-, and 1.7-A resolution, respectively. The
enzyme exists as a tetramer, and the subunit consists of one (alphabeta)8
triose-phosphate isomerase-barrel domain with three functional linkers and one
C-terminal tail. The native enzyme possesses one Zn2+ ion liganded by Glu8,
His10, His164, Asp287, and a water molecule at the active site center, although
the enzyme has been reported to require no cofactor for its catalysis. The
substrate carboxylate takes the place of the water molecule and is coordinated
to the Zn2+ ion. The 2-hydroxy group of the substrate is hydrogen-bonded to
Asp287, which forms a triad together with His218 and Glu221 and is assumed to be
the catalytic base. On the basis of the geometrical consideration, substrate
specificity is uncovered, and the catalytic mechanism is proposed for the novel
Zn2+-dependent decarboxylation.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 3.
FIGURE 3. Schematic diagram showing hydrogen bond and
coordinate bond interactions. Putative hydrogen bond
interactions are shown by dotted lines if the acceptor and donor
are less than 3.5 Å apart. Coordinate bonds between Zn^2+
ion and the active site residues are shown by solid lines if
Zn^2+ ion and the ligand are less than 2.2 Å apart. A,
GRCD in the native form. B, GRCD·2,6-dihydroxybenzoate
complex. C, GRCD·2,3-dihydroxybenzaldehyde complex.
|
 |
Figure 4.
FIGURE 4. Proposed reaction mechanism for decarboxylation
in 2,6-dihydroxybenzoate decarboxylase.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the ASBMB:
J Biol Chem
(2006,
281,
34365-34373)
copyright 2006.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
T.Fujii,
Y.Goda,
M.Yoshida,
T.Oikawa,
and
Y.Hata
(2008).
Crystallization and preliminary X-ray diffraction studies of maleylacetate reductase from Rhizobium sp. strain MTP-10005.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
64,
737-739.
|
 |
|
|
|
|
 |
M.Yoshida,
T.Oikawa,
H.Obata,
K.Abe,
H.Mihara,
and
N.Esaki
(2007).
Biochemical and genetic analysis of the gamma-resorcylate (2,6-dihydroxybenzoate) catabolic pathway in Rhizobium sp. strain MTP-10005: identification and functional analysis of its gene cluster.
|
| |
J Bacteriol,
189,
1573-1581.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
|
 |

